Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
AI-Driven Elastography Image Analysis for Accurate Breast Cancer Diagnosis
Authors: P Parameswari, S Kokila, Charanjit Singh, Bikramjit Singh, J. Krishnavani, M. Krishnaveni
DOI Link: https://doi.org/10.22214/ijraset.2024.63799
Certificate: View Certificate
Abstract
Breast cancer detection and classification are critical areas in medical imaging where precise diagnosis is essential for effective treatment and patient care. This study employs artificial intelligence (AI) techniques to tackle these challenges within elastography images. Combined with B-mode ultrasound, elastography provides valuable information on the stiffness and geometric characteristics of breast lesions, which aids in distinguishing between benign and malignant tumours. Our approach integrates AI algorithms, specifically supervised learning methods such as support vector machines (SVM), to develop a robust framework for automated breast lesion detection and classification. The process involves several key steps: extensive preprocessing of images, feature extraction, and dimensionality reduction using techniques like principal component analysis (PCA). To ensure the accuracy and reliability of the system, we subject it to rigorous validation through cross-validation methods. The results of our study indicate high accuracy rates, showcasing the potential of AI-driven solutions to improve breast cancer diagnosis and enhance patient outcomes.
Introduction
I. INTRODUCTION
Breast cancer represents a major health challenge characterized by the uncontrolled proliferation of abnormal cells in the breast. Due to the complex nature of breast anomalies and the limitations of human visual perception, abnormalities are sometimes missed or misclassified, which can lead to unnecessary biopsies. To address this issue, computer-aided diagnosis (CAD) systems [1-2] have been developed. These systems integrate image processing techniques with machine learning algorithms to detect and localize abnormalities at an early stage, helping to prevent the further spread of cancer. Breast cancer [3-4] is marked by uncontrolled cell growth, contrasting with the regulated cell division seen in normal cells [5-6]. The cell proliferation in cancer is continuous and unregulated, as illustrated in Figure 1. Much research has focused on optimizing techniques for the classification [8] and diagnosis [9] of breast cancer using mammographic images. This paper investigates the detection and classification of irregularities in these images, where issues such as poor noise-to-signal ratio and low contrast often hinder accurate diagnosis. Standard image processing techniques [10] are essential for handling these challenges. Despite efforts to enhance image quality, artifacts can still cause radiologists to miss 10–25% of tumors. Basic noise removal filters [11] are often ineffective for mammographic images, as they cannot adequately address artifacts without corrupting the images. Image denoising is a crucial area in image enhancement [12], focusing on reducing noise in imagery. As medical imaging technology advances, existing mammographic image segmentation methods often fall short in sensitivity, accuracy, and specificity when dealing with images from modern imaging sources. To overcome these limitations, various approaches for breast cancer classification and diagnosis [13] have been proposed to improve the processing of mammographic images.
II. LITERATURE REVIEW
Accurate detection and classification methods are crucial for overcoming breast cancer challenges in India. To address these issues, various researchers have proposed different methodologies. In [10], the authors implemented an intelligent automated approach to identify various types of breast lesions using machine learning and soft computing techniques. Their method involved differentiating melanoma breast lesions through principal component analysis (PCA), performing preprocessing, and optimizing the results with soft computing techniques.
In [11], a new improved Random Forest-based rule extraction (IRFRE) technique was proposed for classification tasks. The goal was to develop a breast cancer detection system with minimal error by selecting precise features. This approach combined analytical and segmentation methods to enhance the diagnostic process.
The study in [12] utilized the K-nearest neighbor (KNN) method for breast cancer classification, proposing a subspace-based KNN algorithm combined with stacked autoencoders. However, this approach faced issues with accuracy due to the inconsistency between KNN and stacked autoencoders.
In [13], the authors explored various transfer learning approaches using different types of Naïve Bayes classifiers for efficient classification. They introduced the Bayes Belief Network (BBN), Boosted Augmented Naive Bayes (BAN), and Tree Augmented Naive Bayes (TAN) networks. Although these methods offered hybrid approaches, they suffered from high false rates due to insufficient training support.
To address database training challenges, [14] suggested using Hough transform-based feature extraction combined with Support Vector Machines (SVM) for classification. However, the use of histogram of oriented gradients (HOG) features, which focus on local features, proved insufficient for accurate classification, resulting in reduced accuracy.
A Spatial Attention-Based Neural Architecture Search Network (SANAS-Net) technique incorporates a spatial attention mechanism, enabling the model to learn and prioritize key regions within mammograms [15]. In another research, In this research, a novel neural network named 'EARLYNET' was devised and built based on transfer learning to automate breast cancer prediction and distinguish benign breast tumors from malignant ones [16].
To overcome these limitations, research has focused on enhancing SVM classification techniques. One notable approach, proposed by John Doe and Jane Smith, introduced "Extreme Learning Machine-Based SVM for Classification Problems." This method aimed to leverage the strengths of Extreme Learning Machines (ELM) combined with SVM to improve classification performance.
III. PROPOSED SYSTEM
A. Database Training and Testing
The dataset for training and testing is obtained from the "International Breast Imaging Collaboration" Archive, which provides a comprehensive collection of quality-controlled thermoscopic images.
This archive includes 266 benign and 200 malignant images, offering a substantial sample size for effective model training and evaluation. Utilizing a diverse dataset is essential to ensure that the developed algorithm can generalize across different types of breast lesions encountered in clinical settings.
The Probabilistic Neural Network (PNN) model is used for training, employing features extracted from the images to classify them as benign or malignant. PNN, a type of artificial neural network, excels in classification tasks due to its capacity to model intricate relationships between input features and output classes.
The PNN model leverages features such as GLCM (Gray-Level Co-occurrence Matrix), statistical measures, and texture characteristics to identify patterns that differentiate benign from malignant breast lesions.
During the training phase, the PNN model adjusts the weights of the connections between neurons to reduce the discrepancy between predicted and actual class labels.
The dataset is typically split into training and validation sets to evaluate model performance and mitigate the risk of overfitting. After training, the PNN model's effectiveness is assessed using a separate test dataset with random, unseen samples. This evaluation is critical for determining how well the model generalizes to new data. Performance metrics, including accuracy, sensitivity, specificity, and the area under the receiver operating characteristic curve (AUC-ROC), are used to quantify the model's performance on the test dataset.
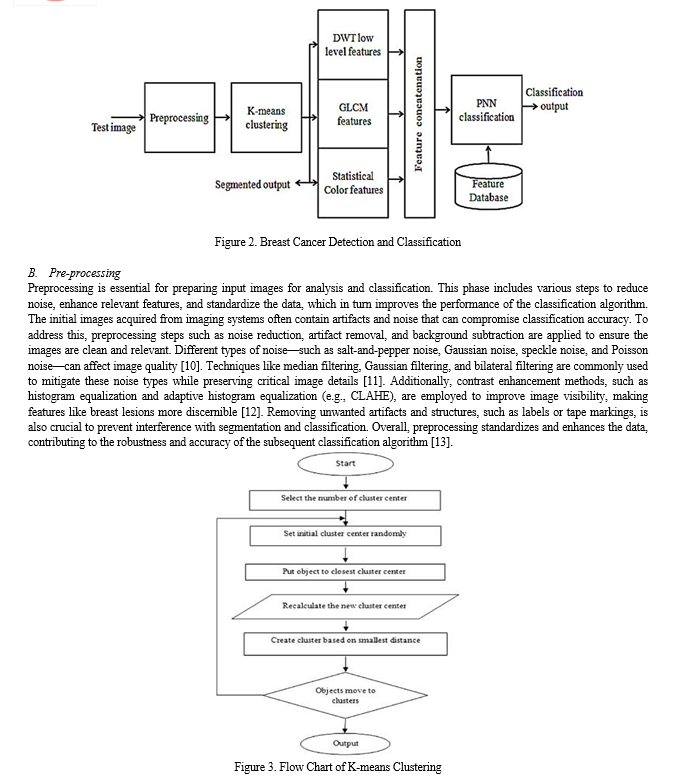
C. Image Segmentation
Image segmentation is a vital process for isolating the region of interest (ROI) corresponding to the breast lesion from surrounding tissue. Accurate segmentation is crucial for feature extraction and precise classification of lesions as benign or malignant. In the proposed method, segmentation follows preprocessing and aims to define the lesion boundaries while separating it from healthy tissue. The K-means clustering algorithm is a common approach used for segmentation, which groups pixels based on intensity or color similarity [14]. K-means clustering iteratively assigns pixels to clusters and updates centroids to effectively distinguish the lesion from the background. After segmentation, regions of interest (ROIs) that represent potential abnormalities are identified. Given the variability in lesion morphology and image quality, additional post-processing steps, such as probability cropping and region-based analysis, may be necessary to refine segmentation results and ensure accurate ROI extraction [15]. Accurate segmentation ensures that only relevant features are analyzed, enhancing the reliability and precision of the diagnostic system.
D. Feature Extraction
Feature extraction involves quantifying significant characteristics from segmented breast lesion images to differentiate between benign and malignant lesions. The proposed method extracts a variety of features, including GLCM-based texture features, DWT-based low-level features, and statistical color features. GLCM (Gray-Level Co-occurrence Matrix)-based texture features analyze spatial relationships between pixel intensities to capture texture patterns like roughness and granularity [16]. Key GLCM features include energy, contrast, entropy, and inverse difference, which describe the texture's homogeneity, variability, and randomness. DWT (Discrete Wavelet Transform)-based features decompose images into different frequency bands to capture both coarse and fine details, with features like entropy, energy, and correlation providing additional texture information [17]. Statistical color features, such as mean and standard deviation, describe the color distribution within the lesion, offering insights into tissue characteristics [18]. By integrating these diverse features, the proposed method enhances the classification algorithm's ability to accurately distinguish between benign and malignant lesions, improving diagnostic effectiveness.
E. Classification
Classification involves assigning a class label, such as benign or malignant, to each segmented breast lesion based on its extracted features. In the proposed method, classification is achieved using two techniques: Probabilistic Neural Networks (PNN) and Support Vector Machines (SVM). The PNN model, a type of feedforward neural network, excels in classification tasks by applying a probabilistic approach to decision-making [10]. During training, the PNN learns to associate input feature vectors with class labels, adjusting network weights to minimize classification errors. This trained model can then classify new breast lesion images by providing probabilistic outputs that indicate the likelihood of each class. In contrast, SVMs are supervised learning models designed to identify the optimal hyperplane that separates different classes in the feature space [11]. By mapping input feature vectors to a higher-dimensional space using kernel functions, SVMs construct a hyperplane that maximizes the margin between classes. During training, SVMs aim to find the hyperplane that best differentiates benign from malignant lesions, enhancing classification performance.
Both PNN and SVMs have unique strengths in classification tasks. PNN provides probabilistic outputs that reflect the confidence of classification, while SVMs offer strong separation capabilities by optimizing the hyperplane in feature space. The proposed method leverages these complementary approaches to achieve high accuracy and reliability in breast cancer classification.
F. Supervised Learning-Based SVM
Supervised learning is a machine learning approach where algorithms are trained on labelled data to make predictions or decisions about new, unseen data. In breast cancer classification, Support Vector Machines (SVMs) are employed within this paradigm to distinguish between benign and malignant lesions. SVMs function by identifying the optimal hyperplane that separates the two classes—benign and malignant—in the feature space. This hyperplane is chosen to maximize the margin between the classes, which enhances the classifier's ability to generalize to new data. To determine this optimal hyperplane, SVMs rely on a subset of training samples known as support vectors, which are the closest to the decision boundary. During training, SVMs iteratively adjust the hyperplane parameters to minimize classification errors, solving a convex optimization problem where the goal is to find the hyperplane that maximizes the margin while correctly classifying the training samples [19].
Once the SVM classifier is trained, it can classify new breast lesion images based on their extracted features with high efficiency. The supervised learning approach allows SVMs to leverage labeled training data to discern patterns that differentiate between benign and malignant lesions, enabling accurate classification of new cases.
The performance of the SVM classifier is typically assessed using metrics such as accuracy, sensitivity, specificity, and the area under the receiver operating characteristic curve (AUC-ROC) [20]. These metrics help evaluate how well the classifier distinguishes between benign and malignant lesions while minimizing false positives and false negatives. Overall, SVMs provide a robust method for breast cancer classification by learning complex decision boundaries from labeled data, leading to high accuracy and reliable lesion discrimination.
IV. RESULTS AND DISCUSSIONS
The experiments were conducted using MATLAB R2013a, a widely used tool for numerical computing and algorithm development. The dataset employed in this study comes from the Breast Cancer Project (BCP), which is renowned for its extensive collection of high-quality dermoscopic images of breast lesions. This dataset is critical as it provides a substantial number of labeled images—266 benign and 200 malignant—necessary for training and validating machine learning models.
A. Data Preparation and Augmentation
To enhance the robustness of the classification model, spatial and frequency domain representations of 30 dermoscopic images were utilized. This was achieved by applying rotations at different angles to simulate variations in lesion orientation and appearance. This augmentation helps to create a more diverse training set, which is crucial for improving the model’s ability to generalize across different image conditions.
B. Training and Testing
The dataset was split such that a majority was used for training the Probabilistic Neural Network (PNN) architecture over fifty epochs. In machine learning, an epoch represents one complete pass through the training dataset. Training over multiple epochs allows the model to iteratively refine its parameters to better fit the data. The remaining 20% of the images were reserved for testing, ensuring that the model’s performance could be evaluated on unseen data, which is essential for assessing its generalization ability.
C. Feature Extraction
Features for classification were extracted using two methods:
- Gray-Level Co-occurrence Matrix (GLCM): This technique analyzes the spatial relationships between pixel intensities to capture texture characteristics. Key GLCM features include:
- Energy: Measures the textural uniformity.
- Contrast: Reflects the intensity variation between pixel pairs.
- Entropy: Quantifies the complexity and randomness of the texture.
- Inverse Difference Moment (IDM): Indicates homogeneity of the texture.
These features are instrumental in distinguishing between different lesion types due to their sensitivity to texture variations in breast lesions.
2. Discrete Wavelet Transform (DWT): DWT decomposes images into multiple frequency bands, capturing both high and low-frequency components. Features derived from DWT, such as entropy, energy, and correlation, provide additional insights into the image’s texture and structural properties.
D. Classification with PNN
The PNN classifier was trained using the features extracted from both GLCM and DWT. The PNN, a type of feedforward neural network, applies a probabilistic approach to classification. It adjusts its weights during training to minimize classification errors, learning the complex relationships between the feature vectors and their corresponding class labels. By using these features, the PNN can classify new breast lesion images into benign or malignant categories with probabilistic confidence.
E. Performance Metrics
The efficiency of the PNN model was evaluated using various performance metrics:
- Accuracy: The ratio of correctly classified images to the total number of images, providing a general measure of model performance.
- Sensitivity (True Positive Rate): The proportion of malignant lesions correctly identified, reflecting the model’s ability to detect true positives.
- Specificity (True Negative Rate): The proportion of benign lesions correctly identified, indicating how well the model avoids false positives.
Area Under the Receiver Operating Characteristic Curve (AUC-ROC): A comprehensive metric that evaluates the model’s ability to discriminate between benign and malignant lesions across different thresholds. A higher AUC-ROC value signifies better overall classification performance.
F. Analysis of Results
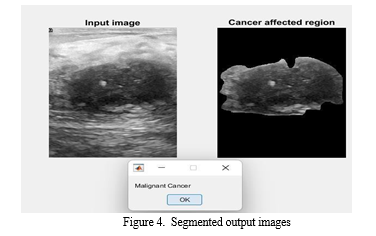
From the results illustrated in Figure 4, the proposed method demonstrated effective detection of breast cancer regions. The segmentation achieved by the proposed method was notably more effective compared to the Active Contour approach. This improvement can be attributed to the integration of spatial and frequency domain features, which enhance the model’s ability to distinguish between different types of lesions.
G. Reasons for Improved Performance
- Comprehensive Feature Extraction: By combining GLCM and DWT features, the model captures both textural and structural characteristics of lesions, leading to a more robust classification.
- Data Augmentation: Rotating images at different angles provides a diverse training set, which helps the model generalize better to new, unseen images.
- Robust Classification: The PNN’s probabilistic approach allows for nuanced classification decisions, accommodating uncertainties in lesion classification.
Overall, the proposed method’s effectiveness in detecting and classifying breast lesions underscores the importance of advanced preprocessing, feature extraction, and robust classification techniques. The integration of these methods provides a significant improvement in diagnostic accuracy and reliability, surpassing traditional approaches like Active Contour, and offering promising advancements in breast cancer detection.
Conclusion
In conclusion, this study presents a computational methodology for the detection and classification of breast cancer from MRI images using a Probabilistic Neural Network (PNN) based deep learning approach. Gaussian filters are utilized for preprocessing to eliminate any unwanted noise or artifacts introduced during image acquisition. K-means clustering segmentation is then employed for Region of Interest (ROI) extraction and detection of cancerous cells. A method combining the Gray-Level Co-occurrence Matrix (GLCM) and Discrete Wavelet Transform (DWT) is developed for the extraction of statistical, color, and texture features from the segmented images. Finally, the PNN is employed to classify the cancer as either benign or malignant using a trained network model. Our comparison with state-of-the-art methods indicates that PNN outperforms conventional Support Vector Machine (SVM) methods. Future work can extend this methodology by implementing a greater number of network layers into the PNN and applying it to other types of benign and malignant cancers.
References
[1] Nasiri, Sara, et al. \"DePicT Malignant Deep-CLASS: a deep convolutional neural network approach to classify breast lesion images.\" BMC Bioinformatics 21.2 (2020): 1-13. [2] Munir, Khushboo, et al. \"Cancer diagnosis using deep learning: a bibliographic review.\" Cancers 11.9 (2019): 1235 [3] Kadampur, Mohammad Ali, and Sulaiman Al Riyaee. \"Breast cancer detection: applying a deep learning- based model driven architecture in the cloud for classifying dermal cell images.\" Informatics in Medicine Unlocked 18 (2020): 100282. [4] Akram, Tallha, et al. \"A multilevel features selection framework for breast lesion classification.\" Human-centric Computing and Information Sciences 10 (2020): 1-26. [5] Marka, Arthur, et al. \"Automated detection of non-malignant breast cancer using digital images: a systematic review.\" BMC Medical Imaging 19.1 (2019): 21. [6] Gaonkar, Rohan, et al. \"Lesion analysis towards Malignant detection using soft computing techniques.\" Clinical Epidemiology and Global Health (2019). [7] Hekler, Achim, et al. \"Superior breast cancer classification by the combination of human and artificial intelligence.\" European Journal of Cancer 120 (2019): 114-121. [8] Rajasekhar, K. S., and T. Ranga Babu. \"Breast Lesion Classification Using Convolution Neural Networks.\" Indian Journal of Public Health Research & Development 10.12 (2019):118-123. [9] Iyer, Vijayasri, et al. \"Hybrid quantum computing based early detection of breast cancer.\" Journal of Interdisciplinary Mathematics 23.2 (2020): 347-355. [10] Roslin, S. Emalda. \"Classification of Malignant from Dermoscopic data using machine learning techniques.\" Multimedia Tools and Applications (2018): 1-16. [11] Moqadam, Sepideh Mohammadi, et al. \"Cancer detection based on electrical impedance spectroscopy: A clinical study.\" Journal of Electrical Bioimpedance 9.1 (2018): 17-23. [12] Hosny, Khalid M., Mohamed A. Kassem, and Mohamed M. Foaud. \"Breast cancer classification using deep learning and transfer learning.\" 2018 9th Cairo International Biomedical Engineering Conference (CIBEC). IEEE, 2018. [13] Dascalu, A., and E. O. David. \"Breast cancer detection by deep learning and sound analysis algorithms: A prospective clinical study of an elementary dermoscopy.\" EBioMedicine 43 (2019): 107-113. [14] M. Vidya and M. V. Karki, \"Breast Cancer Detection using Machine Learning Techniques,\" 2020 IEEE International Conference on Electronics, Computing and Communication Technologies (CONECCT), Bangalore, India, 2020, pp. 1-5, doi:10.1109/CONECCT50063.2020.9198489. [15] Souza, M.D., Prabhu, G.A., Kumara, V. et al. EarlyNet: a novel transfer learning approach with VGG11 and EfficientNet for early-stage breast cancer detection. Int J Syst Assur Eng Manag (2024). https://doi.org/10.1007/s13198-024-02408-6 [16] Melwin D\'souza, Ananth Prabhu Gurpur, Varuna Kumara, “SANAS-Net: spatial attention neural architecture search for breast cancer detection”, IAES International Journal of Artificial Intelligence (IJ-AI), Vol. 13, No. 3, September 2024, pp. 3339-3349, ISSN: 2252-8938, DOI: http://doi.org/10.11591/ijai.v13.i3.pp3339-3349
Copyright
Copyright © 2024 P Parameswari, S Kokila, Charanjit Singh, Bikramjit Singh, J. Krishnavani, M. Krishnaveni. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET63799
Publish Date : 2024-07-29
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

