Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
An Effective Classification of Brain Tumor using Deep Learning Techniques
Authors: Dharshini Sankar Raj, Dr. S. Jeevitha
DOI Link: https://doi.org/10.22214/ijraset.2024.61142
Certificate: View Certificate
Abstract
Brain tumor classification is a critical task in medical imaging, aiding in timely diagnosis and treatment planning. In this paper, we propose a comprehensive approach utilizing deep learning models for the classification of brain tumors into four categories: glioma, meningioma, pituitary tumors, and no tumors. We employ state-of-the-art convolutional neural network (CNN) architectures including ResNet50, DenseNet201, EfficientNetB3, and InceptionV3 for the classification task. To enhance the performance of the model, we employ resizing and augmentation techniques such as flips and rotation, thereby increasing the diversity of the training dataset. This is particularly crucial due to the limitations posed by small-sized datasets in previous methodologies. Our findings underscore the efficacy of deep learning approaches in brain tumor classification, with EfficientNetB3 emerging as a promising model for accurate diagnosis. Furthermore, our utilization of resizing and augmentation techniques demonstrates their significance in mitigating the challenges associated with limited training data.
Introduction
I. INTRODUCTION
Many people are diagnosed with secondary brain tumors, but we don’t know how many exactly. Detecting these tumors early is crucial for effective treatment, but MRI machines sometimes struggle to identify them accurately. This can lead to physical complications and disabilities. Different types of brain tumors exist, and they can affect movement, sight or hearing. Deep learning is a type of artificial intelligence that uses layered computations to teach computers to understand data at multiple levels. This method has improved technologies like speech recognition and object identification. In tumor recognition, deep learning transforms input data into abstract representations, gradually detecting features like tumor edges and shapes automatically.
Supervised learning models are designed to make predictions by learning from labeled datasets. In this process, algorithms receive input data along with corresponding labels to train the model for classification or regression tasks. Deep learning trains models to predict outcomes based on input data or unseen images, with the output impact connected through a single chain of relationships. Unlike parallel exploiters, which create closed-loop chains, none parallel manipulators benefit from additional flexibility and wider workspace. Unsupervised learning is a machine learning technique where the model operates independently without supervision. Instead of providing labeled data, the model learns on its own to discover patterns in unlabeled data. Unlike supervised learning, which relies on labeled data, unsupervised learning allows for more complex processing. It is often considered more exploratory compared to other learning methods.
A. Types Of Brain Tumors
- Pituitary Brain Tumor
Pituitary brain tumor refer to abnormal growth in the pituitary gland. These tumors can affect hormone production, influencing various bodily functions such as growth, organ function, and gland function. While some tumor may lead to hormone overproduction, others result in reduced hormone levels. Most pituitary tumors are benign adenomas, which remain localized and don’t spread to other parts of the body.
2. Glioma Brain Tumor
Glioma originate from the supportive cells surrounding nerve cells, aiding in their function. These tumors can arise from three types of glial cells. Gliomas are classified based on the specific glial cell involved and other inherited characteristics, which can help predict tumor behavior over time and potential treatments. Symptoms include dizziness, headaches, nausea, confusion, memory loss, behavioral changes, urinary problems, vision issues, speech difficulties, and seizures, especially in individuals without a history of seizures.
3. Meningioma Brain Tumor
A meningioma is a tumor that originates from the meninges, the protective membranes surrounding the spinal cord and brain. While not technically a brain tumor, it falls under this category because it can compress nearby brain structures, blood vessels, and nerves. Meningioma is the most common type of tumor found in the head. Symptoms typically develop slowly and may initially be subtle. Depending on the tumor’s location in the brain or spine, symptoms may include changes in vision, headaches (especially worsened in the morning), hearing problems or ringing in the ears, memory loss, seizures, weakness in the arms or legs, and difficulty with language.
II. RELATED WORK
Over the past decade, researchers have increasingly focused on improving brain tumor detection using magnetic resonance images. [1] Traditional approaches typically involve feature extraction from the bottom layer of the network, which may not be optimal for medical images due to their complex nature. To address this limitation, the author proposed a model that utilizes the InceptionV3 convolutional neural network, a deep learning mechanism known for its ability to extract multi-level features. By employing deep learning techniques and optimizing hyperparameters, the model aims to enhance the early detection of brain tumors. In this study, the Adam Optimizer is utilized to optimize the model’s hyperparameters, while a loss function is employed to facilitate the modeling process by guiding the machine to learn from input data. Furthermore, the softmax classifier is employed to classify images into multiple classes, enabling the model to differentiate between various tumor types.
Nyoman and other researchers have explored the use of convolutional neural network (CNNs) for automated tumor classification. They employed a simple CNN architecture consisting of basic layers for convolution, max-pooling, and flattening, followed by a single hidden layer [2]. Despite its simplicity, the CNN achieved impressive results when trained on a dataset of T1 weighted CE-MRI images of glioma, meningioma, and pituitary tumors. The model achieved a training accuracy of 98.51% and a validation accuracy of 84.19%, comparable to more complex region-based segmentation algorithms.
The accurate diagnosis of brain tumor types is critical for effective treatment, and computer-assisted methods have shown promise in improving diagnostic accuracy. In this study [3], a novel approach using a convolutional neural network based on complex networks with a modified activation function is proposed for the classification of brain tumors from magnetic resonance imaging (MRI) scans. The results of the modified model demonstrate its effectiveness in brain tumor classification, achieving an impressive accuracy of 95.49%. Furthermore, the model outperforms several established models such as ResNet, DenseNet, indicating its superior performance in tasks, J.S. Paul and his researchers study aims to leverage deep learning methods for the classification of different types of brain tumors, including meningioma, glioma, and pituitary tumors. The research dataset comprises a substantial collection of T1-weighted contrast-enhanced MRI (CE-MRI) brain images, totaling 3064 images from 233 patients across axial, coronal, and sagittal planes. To ensure clarity and avoid confusion, the study [4] focuses specifically on the axial images, consisting of 989 images from 191 patients. Further experimentation involves augmenting the original axial images to enhance the training process. The findings of the study reveal that training neural networks on axial data yields highly accurate classifications, with an average five-fold cross-validation accuracy of 91.43% achieved by the best-trained neural network.
The field of medical image analysis, particularly in computer-aided diagnosis (CAD) for brain tumor classification, has witnessed a surge in research focusing on deep learning techniques. [5] One notable approach involves utilizing pre-trained convolutional neural networks (CNNs) and transfer learning to extract meaningful features from brain MRI images for accurate tumor classification. Studies have explored architectures like GoogleNet, VGGNet, and ResNet, assessing their performance in differentiating among glioma, meningioma, and pituitary tumors. Comparatively analyses have been conducted to evaluate the effectiveness and efficiency of these models, often employing patient-level cross validation techniques and standardized evaluation metrics. Results consistently indicate promising classification accuracies, with some studies reporting mean accuracies of up to 98%, outperforming traditional methods.
Advancements in technology have led to the utilization of 3D scanning for tumor analysis, as discussed in [6]. The study explores 3D image processing methods for brain tumor detection and classification, employing deep learning frameworks like MobileNetV2, MobileNetV3 (both small and large variants), VGG16, VGG19, and custom CNN models. Among these, CNNs demonstrated superior accuracy, highlighting their effectiveness in tumor analysis. The proposed solution integrates a CNN developed with keras and TensorFlow with a comprehensive cross-platform application built using PYQt5 and MariaDB. This integrated approach is tailored for medical environments such as hospitals, enabling the analysis of clinical images. The primary objective of the research is to characterize brain damage caused by tumors using real-world data and detect abnormal pixels [7].
III. DEEP LEARNING METHODS
Dataset preparation, pre-processing, model training, and categorization are the four key phases of deep learning techniques used to diagnose brain tumors.
A. Dataset Preparation
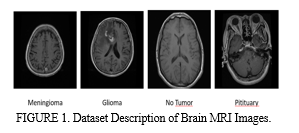
The dataset mentioned in [8] is publicly accessible and utilized for assessing the performance of classification and retrieval algorithms. Comprising 3265 brain MRI images, the dataset encompasses instances diagnosed with one of three tumor types: meningioma, glioma, and pituitary tumors, along with images indicating the absence of tumors. These images are captured using the T1-CE MRI modality and are presented in coronal, sagittal, and axial views. Specifically, the dataset comprises 926 brain MRI images depicting glioma, 937 images representing meningioma cases, and 902 images corresponding to instance of pituitary tumors. Additionally, 500 images are included in the dataset that don’t exhibit any tumors. The images are stored in .jpg format having dimension of either 512x512 or 254x254 pixels. This dataset provides a valuable resource for researchers and practitioners in the field, facilitating the development and evaluation of algorithms aimed at brain tumor classification. Table 1 and Figure 1 demonstrates the dataset description.
|
Glioma |
926 images |
|
Meningioma |
937 images |
|
No Tumor |
500 images |
|
Pituitary |
902 images |
TABLE 1. Description of Brain MRI Images by Tumor Type.
B. Data Preprocessing
- Resizing
In the process of preparing the dataset of the research, we undertook crucial steps in data preprocessing, particularly in resizing the images to ensure uniformity and compatibility for subsequent analysis. Resizing refers to the adjustment of image dimensions to a standardized size, facilitating uniform processing and analysis across the dataset. Specifically, we resized the images to a standard dimension of 160x160 pixels, a common practice in image processing tasks. However, it’s worth noting that in some studies [9], images are resized to 256x256 pixels, which may offer increased detail and resolution for analysis. Through python programming, specifically employing the OpenCV library, we iteratively processed the images from both the training and testing folders. Each image underwent resizing using the cv.resize() function, which allowed us to adjust the dimensions uniformly across the dataset. This meticulous data preprocessing lays the foundation for the subsequent phases of the model, enabling to leverage advanced machine learning techniques for effective brain tumor classification and diagnosis.
2. Data Augmentation
Data augmentation is a fundamental strategy in machine learning and computer vision, particularly pivotal in scenarios where dataset size or diversity is constrained. This technique involves the creation of additional training data by applying diverse transformations to existing samples, such as rotations, translations, flips, zooms, and color variations. The objective is to augment the diversity [10] of the training dataset, thereby bolstering the robustness and generalization capacity of machine learning models.
The data augmentation is implemented utilizing the Image Data Generator from the keras module, enabling real-time augmentation during model training. Notably, augmentation parameters like rotation range, width, and height shift ranges, zoom range, and horizontal flips are tailored to introduce variations in the training images.
Figure 2 demonstrates the augmentation process done in the model. Furthermore, to mitigate storage requirements and optimize computational resources, the augmentation process is performed on-the-fly. This on-the-fly augmentation approach generated augmented samples dynamically during model training, eliminating the need to store the augmented data separately. This not the only conserves the storage space [11] but also streamlines the training process by generating augmented samples in real-time, enhancing efficiency and scalability. Data augmentation stands as a pivotal technique [25] for enhancing the performance and robustness of machine learning models, particularly in scenarios constrained by limited training data. Before data augmentation, the training set comprised 3265 images with dimension of 160x160 pixels and 3 color channels, while the testing set consisted of 3265 samples. Following augmentation, the training set expanded significantly to 9730 images, maintaining the same dimensions and color channels. Likewise, the testing set also expanded to 9730 samples. Upon closer inspection, it is evident that the augmentation process balanced the distribution of tumor categories within both the training and testing sets. Table 2 shows the detailed version of data augmentation. The training data exhibited varying counts across tumor categories, with 2349 samples for category 0, 2371 for category 1, 1269 for category 2, and 2281 for category 3. Similarly, the testing data displayed similar disparities, with 414, 419, 224, and 403 samples for categories 0, 1, 2, and 3, respectively.
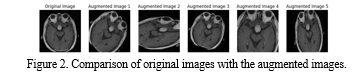
Post-augmentation, the training and testing sets showcased a more equitable distribution across tumor categories, with each category now comprising approximately one-fourth of the dataset. This balanced representation ensures that the model is exposed to a diverse range of tumor types during training and evaluation, enhancing its ability to generalize effectively to unseen data and improving overall performance and reliability.
The dataset has been divided into two distinct subsets: Training and Testing. This division is crucial for evaluating the performance of machine learning models. The split is accomplished using the test size parameter, which determines the proportion of data allocated to each set. In this particular scenario, a test size of 15% has been employed, signifying that 15% of the dataset is reserved for testing purposes, while the remaining 85% is designated for training the model. This partitioning strategy ensures that the model is trained on a substantial portion of the dataset, allowing it to learn the underlying patterns and relationships present in the data. Meanwhile, the testing set serves as an independent sample for assessing the model’s generalization capability and performance on unseen data. By segregating the dataset into training and testing subsets, we can effectively gauge the model’s predictive accuracy and identify any potential issues such as overfitting or underfitting. This systematic approach to data splitting facilitates robust model evaluation and enhances the reliability of the machine learning process.
|
Tumor Types |
Before Augmentation |
After Augmentation |
||
|
Training |
Testing |
Training |
Testing |
|
|
Glioma |
787 |
139 |
2350 |
415 |
|
Meningioma |
796 |
141 |
2373 |
419 |
|
No Tumor |
425 |
75 |
1266 |
223 |
|
Pituitary |
767 |
135 |
2281 |
403 |
Table 2. Tumor type distribution before and after data augmentation.
C. Model Training
Model training involves the process of feeding labeled data into a machine learning model, allowing it to learn patterns and relationships between input features and output labels. In the context of brain tumor classification, we utilize pre-trained convolutional neural network (CNN) architectures such as ResNet50, DenseNet201, EfficientNetB3, and InceptionV3. These models, available in keras library, are initialized with weights learned from large datasets like ImageNet, enabling them to capture intricate features from input images effectively.
Additionally, data augmentation techniques are applied to mitigate overfitting, a common issue when dealing with limited datasets. Through this approach, we aim to train robust models capable of accurately classifying brain tumor types, enhancing diagnostic capabilities in medical applications.
- Pre-trained Models
a. ResNet50
ResNet50 introduced residual learning, which simplifies the deduction of input characteristics [21] from specific layers. This is achieved through shortcut connections between every pair of the thirty-three filters, directly linking the input of the kth layer to the (k+x)th layer. This prevents vanishing gradients by reusing initializations from the preceding layer until the adjacent layer has learned its weights.
b. DenseNet201
DenseNet201 is a convolutional neural network with a depth of 201 layers. It offers a pretrained version trained on over a million images from the ImageNet database. This pretrained network has the capability to classify images into 1000 object categories, including various objects and animals.
c. EfficientNetB3
EfficientNet is a convolutional neural network architecture and scaling approach that adjusts all dimensions of depth, width, and resolution using a compound coefficient. Unlike traditional methods that independently scale these factors, [23] EfficientNet uniformly scales network width, depth, and resolution using predetermined coefficients. This scaling method is based on the idea that larger input images require additional layers to enhance the receptive field and more channels to capture finer details.
d. Inceptionv3
InceptionV3, a popular CNN architecture, consists of eleven stacked inception models. Each model includes pooling layers and convolutional filters with rectified linear units as the activation function. The model takes a two-dimensional image input comprising sixteen brain slices arranged in a 4-3-4 grid after preprocessing. This pre-trained model was trained on the ImageNet dataset and fine-tuned with a batch size and a learning rate of 0.0001.
e. Classifier Settings
In the process of training our models for brain tumor classification, we employed various pre-trained convolutional neural network architectures, including EffectiveNetB3, DenseNet201, ResNet50, and InceptionV3. Each model was initialized with weights obtained from ImageNet and adapted to classify brain tumor types based on extracted features from MRI images. To enhance model performance and mitigate overfitting, we utilized a range of hyper parameters and optimization techniques, such as dropout regularization and the Adam optimizer. Training was conducted on a dataset comprising labeled brain MRI images, with the dataset divided into training and testing sets to evaluate model generalization. The training process involving iteratively feeding batches of data into the models, adjusting the model weights based on computed loss, and optimizing performance metrics like accuracy.
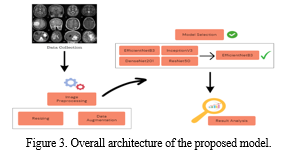
For the proposed framework using EfficientNet, we developed a custom model architecture by integrating the EfficientNet base model with additional layers for classification. Figure 3 shows the architecture of proposed methodology. The architecture included global average pooling layers, dropout regularization, and fully connected dense layers with appropriate activation functions. During training, the model achieved exceptional accuracy of 99% on the training set and 97% on the testing set, demonstrating robust performance in classifying the brain tumor types. By leveraging EfficientNet’s efficient scaling technique and feature extraction capabilities, our framework achieved superior classification accuracy while maintaining computational efficiency, making it suitable for deployment in real-world medical applications. The EfficientNet based model offers a promising approach for accurate and efficient brain tumor classification, contributing to advancements in computer-aided diagnosis and improving patient care outcomes.
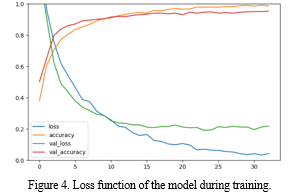
The EfficientNet was trained using the training set after the preprocessing. Figure 4 shows the hyperparameters of the network that were heuristically adjusted so as to facilitate the convergence of the loss function during training. The model architecture is being trained for a specific task, likely related to image classification given the use of softmax activation in the final layer. The choice of Adam optimizer with a specific set of hyper parameters indicated a thoughtful consideration of the learning dynamics during training. The dropout layers incorporated in the model architecture with the dropout rates of 0.55 and 0.3 suggests an approach to regularization, aimed at reducing the model’s reliance on specific features during training to enhance the generalization performance. The choice of GlorotNormal initializer for the dense layers emphasizes the importance of initializing model parameters appropriately to facilitate effective training. Table 3 shows the hyperparameters settings of the model.
|
Optimizer |
Adam |
|
Learning Rate |
0.000016 |
|
Loss |
Sparse Categorical |
|
Activation Function |
Softmax |
|
Kernel Initializer |
GlorotNormal |
|
Epoch |
90 |
Table 3. Experimental Parameters.
2. Performance Metrics and Evaluation
Various metrics are utilized to access the performance of a classifier, with classification accuracy being the most commonly used index. It quantifies the proportion of correctly classified samples relative to the total number of data samples. In our experiments, the EfficientNetB3 model achieved the highest accuracy at 97%, outperforming DenseNet201, ResNet50, and InceptionV3, which attained accuracies of 95% and 94%, respectively. The results highlight the superior performance of EfficientNetB3 in brain tumor classification. However, accuracy alone may not suffice when dealing with imbalanced datasets, as is the case in our classification problem. Therefore, additional performance metrics are necessary for a comprehensive evaluation. Confusion matrices are employed to analyze the classifier’s performance, enabling the derivation of metrics such as precision, recall, and specificity for each tumor class. Notably, the specificity values are high across all classes, indicating accurate identification of samples without a particular disease. The F-score, calculated as the harmonic mean of precision and recall, provides a comprehensive measure of classification performance, particularly beneficial in the presence of class imbalances. Table 4 presents the category-specific performance metrics obtained when employing the EfficientNetB3 with the features.
|
Tumor Type |
Precision |
Recall |
F1-score |
|
Meningioma |
0.96 |
0.97 |
0.96 |
|
Pituitary |
0.96 |
0.96 |
0.96 |
|
Glioma |
0.97 |
0.99 |
0.98 |
|
No Tumor |
0.99 |
0.98 |
0.99 |
TABLE 4. Class-Specific evaluation of brain tumor classifier.
D. Comparison With Related Work
In the realm of brain tumor classification using MRI, several models have been developed to tackle this challenging tasks. One notable model, referred to as Brain Tumor MRI classification, employed the InceptionV3 algorithm with an Adam optimizer. This model [13] was trained on a dataset comprising of 2880 samples, with 800 allocated for testing and an additional 800 for validation. Impressively, it achieved commendable accuracy rates of 99.34% on the training set and 94% on the testing set.
Another noteworthy model, [12] adopted a CNN-based approach and also utilized the Adam optimizer with cross-entropy loss. Trained on a dataset consisting of 1,672 samples, with 207 reserved for testing and 186 for validation, this model achieved an accuracy of 94% on the training set and 89% on the testing set. In comparison to these existing models, our proposed model harnesses the power of EfficientNetB3, a cutting-edge neural network architecture known for its superior performance in image classification tasks. Leveraging the Adam optimizer and sparse categorical cross-entropy loss, our model was trained on a sizable dataset comprising 2880 samples, with 800 samples for testing and an additional 800 for validation. Impressively our model achieved outstanding accuracy rates of 99.32% on the training set and 97.14% on the testing set. Table 4 shows the comparison of the existing work with the proposed model. While the existing models have demonstrated notable performance in brain tumor classification, our model stands out for its exceptional accuracy rates on both the training and testing sets. The utilization of EfficientNetB3, coupled with meticulous training and optimization, has enabled our model to
|
Title |
Dataset |
Algorithm |
Loss |
Optimizer |
Accuracy |
|||
|
An Effective Classification of Brain Tumor using Deep Learning Techniques [Proposed] |
Training |
8270 |
EfficientNetB3 |
Sparse Categorical Cross Entropy |
Adam |
Training |
99.32% |
|
|
Testing |
1460 |
Testing |
97.14% |
|||||
|
Brain tumor magnetic resonance image classification: a deep learning approach |
Training |
2880 |
InceptioZn V3 |
Categorical Cross Entropy |
Adam |
Training |
99.34% |
|
|
Testing |
800
|
|||||||
|
Testing |
94% |
|||||||
|
Validation |
||||||||
|
800 |
||||||||
|
Brain tumor detection from MRI images using deep learning techniques |
Training |
1672 |
CNN |
Cross Entropy |
Adam |
Training |
94% |
|
|
Testing |
207 |
|||||||
|
Testing |
89% |
|||||||
|
Validation |
186 |
|||||||
Table 4. Comparison of existing works with proposed model.
achieve remarkable results, showcasing its potential for real-world application in medical imaging. Furthermore, the robustness of our model’s performance underscores its effectiveness in accurately detecting and classifying brain tumors from MRI images, thereby offering valuable insights for clinical diagnosis and treatment planning.
Conclusion
In conclusion, our study presents a novel approach to brain tumor classification utilizing the EfficientNetB3 neural network architecture. With impressive accuracy of 99.32% on the training set and 97.14% on the testing set, our model demonstrated significant advancements in the field of medical image analysis. By leveraging state-of-the art deep learning techniques and optimizing model parameters, we have achieved superior performance compared to existing models, thereby enhancing the reliability and accuracy of brain tumor classification from MRI images. There are several avenues for future research and development in this domain. Firstly, further exploration of advanced neural network architectures, such as transformer-based models, could potentially improve classification accuracy and computational efficiency. Additionally, incorporating multi-modal imaging data, including functional MRI and diffusion tensor imaging, may provide richer information for more comprehensive tumor characterization. Furthermore, efforts to address class imbalance in the dataset through techniques like data augmentation and class weighting may lead to robust and generalizable models. Overall, our research represents a significant step forward in the field of brain tumor classification using deep learning techniques. By continuing to innovate and collaborate across disciplines, we can harness the full potential of artificial intelligence to improve diagnostic accuracy, treatment planning, and patient outcomes in the field of neuroimaging.
References
[1] M. J. Lakshmi and S. N. Rao, ‘‘Brain tumor magnetic resonance image classification: A deep learning approach,’’ Soft Comput., vol. 26, no. 13, pp. 6245–6253, Jul. 2022, doi: 10.1007/s00500-022-07163-z [2] N. Abiwinanda, M. Hanif, S. T. Hesaputra, A. Handayani, and T. R. Mengko, ‘‘Brain tumor classification using convolutional neural network,’’ in Proc. World Congr. Med. Phys. Biomed. Eng., vol. 68, 2018, pp. 183–189, doi: 10.1007/978-981-10-9035-6_33. [3] Z. Huang, X. Du, L. Chen, Y. Li, M. Liu, Y. Chou, and L. Jin, ‘‘Convolu tional neural network based on complex networks for brain tumor image classification with a modified activation function,’’ IEEE Access, vol. 8, pp. 89281–89290, 2020, doi: 10.1109/ACCESS.2020.2993618. [4] J. S. Paul, A. J. Plassard, B. A. Landman, and D. Fabbri, ‘‘Deep learn ing for brain tumor classification,’’ in Proc. SPIE, vol. 10137, 2017, pp. 1013710–1013726, doi: 10.1117/12.2254195. [5] S.DeepakandP.M.Ameer,‘‘Brain tumor classification using deep CNN features via transfer learning,’’ Comput. Biol. Med., vol. 111, pp. 1–7, Aug. 2019, doi: 10.1016/j.compbiomed.2019.103345. [6] M. A. H. Tuhin, T. Pramanick, H. K. Emon, W. Rahman, M. M. I. Rahi, and M. A. Alam, ‘‘Detection and 3D visualization of brain tumor using deep learning and polynomial interpolation,’’ in Proc. IEEE Asia Pacific Conf. Comput. Sci. Data Eng. (CSDE), Dec. 2020, pp. 1–6, doi: 10.1109/CSDE50874.2020.9411595. [7] S. Pokhrel, L. K. Dahal, N. Gupta, R. Shrestha, A. Srivastava, and A. Bhasney, ‘‘Brain tumor detection application based on convolutional neural network,’’ in Proc. 2nd Int. Conf. Intell. Technol. (CONIT), Jun. 2022, pp. 1–5, doi: 10.1109/CONIT55038.2022.9848177. [8] https://github.com/deeptangshu29/brain-tumor-detection/tree/main/data Dataset Description Link. [9] Pashaei A., Sajedi H., Jazayeri N.: Brain tumor classification via convolutional neural network and extreme learning machines.IEEE 8th International Conference on Computer and Knowledge Engineering.2018.ICCKEpp. 314-319. [10] Xue D.X., Zhang R., Feng H., Wang Y.L.: CNN-SVM for microvascular morphological type recognition with data augmentation. J. Med. Biol. Eng. 2016; 36: pp. 755-764. [11] Goeric Huybrechts, Thomas Merritt, Giulia Comini, Bartek Perz, Raahil Shah and Jaime Lorenzo-Trueba, \"Low-resource expressive text-to-speech using data augmentation\", ICASSP 2021–2021 IEEE International Conference on Acoustics Speech and Signal Processing (ICASSP), pp. 6593-6597, 2021. [12] M. J. Lakshmi and S. N. Rao, ‘‘Brain tumor magnetic resonance image classification: A deep learning approach,’’ Soft Comput., vol. 26, no. 13, pp. 6245–6253, Jul. 2022, doi: 10.1007/s00500-022-07163-z. [13] P. G. Brindha, M. Kavinraj, P. Manivasakam, and P. Prasanth, ‘‘Brain tumor detection from MRI images using deep learning techniques,’’ Mater. Sci. Eng., vol. 1055, pp. 1–8, Feb. 2021, doi: 10.1088/1757-899X/1055/1/012115. [14] S. Zhang and G. Xu, \"A novel approach for brain tumor detection using MRI Images\", Journal of Biomedical Science and Engineering, vol. 9, no. 10, pp. 44-52, 2016. [15] D. Lamrani, B. Cherradi, O. El Gannour, M. A. Bouqentar and L. Bahatti, \"Brain tumor detection using mri images and convolutional neural network\", International Journal of Advanced Computer Science and Applications, vol. 13, no. 7, 2022. [16] H. H. Sultan, N. M. Salem and W. Al-Atabany, \"Multi-classification of brain tumor images using deep neural network\", IEEE access, vol. 7, pp. 69215-69225, 2019. [17] T. Sadad, A. Rehman, A. Munir, T. Saba, U. Tariq, N. Ayesha, et al., \"Brain tumor detection and multi-classification using advanced deep learning techniques\", Microscopy Research and Technique, vol. 84, no. 6, pp. 1296-1308, 2021. [18] M. M. Badža and M. ?. Barjaktarovi?, \"Classification of brain tumors from mri images using a convolutional neural network\", Applied Sciences, vol. 10, no. 6, pp. 1999, 2020. [19] M. A. Hamid and N. A. Khan, \"Investigation and classification of mri brain tumors using feature extraction technique\", Journal of Medical and Biological Engineering, vol. 40, pp. 307-317, 2020. [20] M. Sajjad, S. Khan, K. Muhammad, W. Wu, A. Ullah and S. W. Baik, \"Multi-grade brain tumor classification using deep CNN with extensive data augmentation\", Journal of computational science, vol. 30, pp. 174-182, 2019. [21] Diakogiannis, Foivos Waldner, Francois Caccetta, Peter Wu and Chen, \"ResUNet-a: A deep learning framework for semantic segmentation of remotely sensed data\", ISPRS Journal of Photogrammetry and Remote Sensing, vol. 16, pp. 94-114, 2020. [22] SA. Shenbagarajan, V. Ramalingam, C. Balasubramanian and S. Palanivel, \"Tumor Diagnosis in MRI Brain Image using ACM Segmentation and ANN-LM Classification Techniques\", Indian Journal of Science and Technology, vol. 9, no. 1, January 2016. [23] Maria Nazir, Sadia Shakil and Khurram Khurshid, \"Role of deep learning in brain tumor detection and classification (2015 to 2020): A review\", Computerized Medical Imaging and Graphics, vol. 91, pp. 101940, 2021. [24] Sarah Ali Abdelaziz Ismael, Ammar Mohammed and Hesham Hefny, \"An enhanced deep learning approach for brain cancer MRI images classification using residual networks\", Artificial intelligence in medicine, vol. 102, pp. 101779, 2020. [25] Adrian Rosebrock, Keras ImageDataGenerator and Data Augmentation, 2019, [online] Available: www.pyimagesearch.com.
Copyright
Copyright © 2024 Dharshini Sankar Raj, Dr. S. Jeevitha. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET61142
Publish Date : 2024-04-27
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

