Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Bio-Medical Image Segmentation And Detection For Brain Tumour And Skin Lesions Diseases Through U-NET
Authors: Sailaja Y, Suresh Kumar Gummadi, Harshitha Gorrepati, Deepika Pucha
DOI Link: https://doi.org/10.22214/ijraset.2024.59064
Certificate: View Certificate
Abstract
The research concentrates on enhancing the precise identification and localization of diseases within medical images, a pivotal component of medical imaging analysis crucial for diagnoses and treatment planning. Employing the U-Net architecture, renowned for its effectiveness in biomedical image segmentation, the study targets the detection of brain tumors and skin lesions in MRI or CT scans. Remarkably, the proposed method not only identifies diseases but also provides intricate details regarding their dimensions and spatial arrangement. Extensive experimental validation showcases the method\'s superiority over existing approaches, underscoring its potential for holistic learning in medical image analysis.
Introduction
I. INTRODUCTION
As biomedical technology progresses, diverse sectors are harnessing biomedical signals from imaging for a multitude of applications, resulting in a significant increase in image generation. These images play a pivotal role in aiding healthcare practitioners in promptly pinpointing anatomical abnormalities and monitoring disease evolution accurately. Yet, the sheer abundance of image data presents a challenge, particularly with a scarcity of experienced medical professionals. Relying solely on manual processing would be impractical, consuming substantial time and labor and leading to notable inefficiencies.
The utilization of the U-Net architecture in the Bio-Medical Image Segmentation System marks a significant advancement in the realm of medical image analysis. Tailored for semantic segmentation tasks, U-Net stands out for its adeptness at capturing intricate details and structures within biomedical images. Its unique structure includes a pathway for capturing contextual information and another for precise localization using high-resolution feature maps. In the domain of biomedical image segmentation, U-Net shines in accurately delineating challenging structures such as brain tumors or skin lesions from medical images. complex structures like brain tumors or skin lesions from medical images.
II. LITERATURE SURVEY
In his research, Ramin Ranjbarzadeh conducted a review aimed at developing a flexible and efficient brain tumor segmentation system. The thesis of his work proposed a preprocessing approach focused on analyzing only a small portion of the image rather than the entire image. To capture diverse tumor features effectively, Ranjbarzadeh employed four modalities: fluid attenuated inversion recovery (FLAIR), T1-contrasted (T1C), T1-weighted (T1), and T2-weighted (T2) images as input. Additionally, corresponding Z-Score normalized images of these modalities were utilized to enhance the segmentation results' Dice score without introducing complexity to the model's architecture. Ranjbarzadeh introduced a cascade CNN model that integrates both local and global data extracted from diverse sources MRI modalities. Furthermore, he proposed a distance-wise attention mechanism to account for the brain tumor's location across the four input modalities. For skin lesion segmentation, the implementation relied on TensorFlow. The RATS 2018 dataset served as the basis for brain tumor analysis in his review.
Lina Liu's investigation aimed to devise an adaptable and effective framework for segmenting skin lesions. They utilized a convolutional neural network (CNN) with a Pyramid Pooling Module (PPM) as the main structure. To refine segmentation precision, they incorporated an additional edge prediction task by employing parallel branches interconnected with Cross-Connection Layer (CCL) modules. Additionally, they incorporated a Multi-Scale Feature Aggregation (MSFA) module to extract information from various scale feature maps. The final outcome was produced via PyTorch implementation, where predictions were amalgamated using a weighted sum determined by 1x1 convolution. Evaluation utilized the SBI2017 dataset for analysing skin lesions.
III. EXISTING SYSTEM
Provided two existing systems for Brain Tumour and skin lesions.
A. Brain Tumour
The current approach to segmenting brain tumours involves the utilization pertaining to deep neural networks techniques coupled with an attention mechanism applied to multi-modal MRI images. By incorporating various MRI modalities such as FLAIR, T1C, T1, and T2, the approach aims to gather comprehensive insights into both the anatomy and pathology of the brain. The attention mechanism enables the model to focus on pertinent regions within the images, thereby enhancing segmentation precision. At the heart of this approach lies a convolutional neural network (CNN) architecture specifically tailored for efficiently processing multi-modal MRI data. Moreover, advanced techniques for feature fusion are implemented to amalgamate information from diverse modalities, thereby enhancing the model's capacity to capture a wide range of tumour characteristics. Overall, the current system approach strives to achieve precise and resilient segmentation of brain tumours, which is pivotal for clinical diagnosis and treatment planning.
While this approach boasts several strengths, it faces a potential drawback concerning its limited generalization, especially when encountering unfamiliar datasets or diverse patient demographics. If the model undergoes training on a dataset with limited variability, it may encounter challenges in accurately segmenting tumours across different imaging conditions or varied clinical contexts. Overcoming this limitation is essential to uphold the approach's efficacy and relevance in real-world clinical applications.
???????B. Skin Lesions
The current system emphasizes skin lesion segmentation through the utilization of deep learning techniques, integrating an auxiliary task to improve performance. This likely involves the development of a customized deep learning architecture, potentially a convolutional neural network (CNN), specifically tailored for the task of segmenting skin lesions. The inclusion of an auxiliary task alongside the primary segmentation objective aims to enhance the model's accuracy by leveraging additional information or regularization techniques. Model training involves annotated datasets of skin lesion images paired with corresponding segmentation masks, with evaluation conducted using standard metrics such as the Dice similarity coefficient or Intersection over Union (IOU). Overall, this approach strives to advance skin lesion segmentation accuracy through deep learning methodologies while addressing challenges such as limited generalization and data availability. Obtaining diverse and extensive datasets with annotated skin lesion images for training presents a notable obstacle. The limited availability of high-quality annotated data might impede the model's ability to adjust to different types of skin lesions and imaging conditions.
IV. PROPOSED METHOD
Automating the detection of brain tumours and skin lesions is vital for early diagnosis and improving patient outcomes. Manual detection methods are time-consuming, but integrating the U-Net Sophisticated machine learning approach that excels in discerning complex patterns through neural network architectures can provide valuable assistance to both physicians and patients. Our application focuses on predicting brain tumour locations using publicly accessible MRI datasets from The Cancer Imaging Archive (TCIA). These datasets contain manually annotated FLAIR abnormality segmentation masks from 110 patients in The Cancer Genome Atlas (TCGA) lower-grade glioma collection. Each image, sized at 256x256 pixels, merges MRI slices from three modalities into RGB format. Furthermore, we leverage an additional dataset for skin lesion classification, combining data from HAM10000 (2019) and MSLDv2.0, encompassing 14 different types of skin lesions.
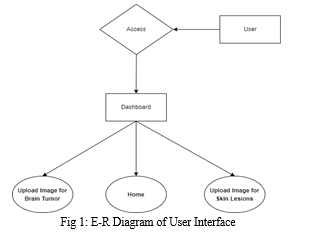
???????A. User Interface
The system's user interface is tailored to cater to two distinct medical conditions: brain tumors and skin lesions. Users engage with the system by uploading medical images pertinent to these conditions. The system's output comprises masked images that pinpoint and isolate the identified areas of interest, streamlining the process for in-depth analysis.
The foundation of this system for biomedical image segmentation relies on the U-Net model, a convolutional neural network architecture recognized for its excellence in semantic segmentation tasks. U-Net's standout feature lies in its remarkable precision in delineating complex structures within medical images, making it a preferred choice for tasks such as identifying brain tumors and skin lesions. The system utilizes Django framework for web development, employing HTML, CSS, and JavaScript for front-end functionalities, while relying on an MSSQL database for efficient data management. Python powers the back-end operations, facilitating the processing of images. This configuration seamlessly integrates the U-Net model into a user-friendly web application designed for medical image segmentation.
In this project, the front end makes use of HTML, CSS, and JavaScript, while Python is employed for the back end. The system's functionality is backed by the Django framework and a MySQL Database, providing a robust infrastructure.
B. ?U-NET Implementation
U-Net stands out as a specialized architecture tailored specifically for semantic segmentation tasks. Its name originates from its unique architectural design resembling a "U." This model comprises convolutional layers intricately connected in two networks. Its architecture features a contracting path, dedicated to capturing contextual information, and an expansive path, focused on refining spatial details. This distinctive layout enables the efficient integration of both local and global features from the input data, facilitating robust segmentation.
We've crafted our implementation of the U-Net architecture utilizing Python alongside prevalent deep learning libraries. Within our system, we input RGB MRI images depicting brain tumours or RGB images depicting skin lesions, each containing 3 channels. Our aim isn't to classify these images but rather to produce a mask identical in size to the input image.
To achieve our goal, we utilize an encoder network, also referred to as the contracting network. Its main purpose is to reduce the spatial dimensions of the input while increasing the number of channels. This network comprises four encoder blocks, each composed of two convolutional layers with a 3x3 kernel size and valid padding, followed by a Rectified Linear Unit (ReLU) activation function. Afterward, the output from each block undergoes a max-pooling layer with a 2x2 kernel size and a stride of 2. This max-pooling operation effectively decreases the spatial dimensions, aiding in reducing computational costs during training. The final output of the encoder layer is an image with dimensions 16x16x1024, representing a feature map of the input image. Convolutional operations within the encoder are depicted by blue arrows, while max-pooling operations are indicated by red arrows in the model architecture. This architectural layout facilitates the extraction of hierarchical features from the input images, preparing them for subsequent processing in the decoder section of the U-Net architecture.
Situated between the encoder and decoder networks, the bottleneck layer occupies a central position within the architecture. Comprising two convolutional layers followed by Rectified Linear Unit (ReLU) activation, this layer serves as the final representation of the feature map.
The decoder network, also termed the expansive network, operates in opposition to the encoder, expanding spatial dimensions while reducing channel numbers. This entails increasing the size of the feature maps to match the input image's dimensions, as depicted by green arrows in the architectural diagram. By leveraging skip connections, the decoder network receives the feature map from the bottleneck layer and generates a segmentation mask. It consists of four decoder blocks, initiating with a transpose convolution using a 2x2 kernel size and stride of 2. The resultant output is fused with the corresponding skip layer connection from the encoder block. Subsequently, two convolutional layers employing a 3x3 kernel size are applied, followed by a ReLU activation function, ultimately producing an image with dimensions 256x256x64.
Skip connections, illustrated as black arrows in the architectural diagram, play a crucial role in leveraging contextual feature information from encoder blocks to generate the segmentation map. These connections combine high-resolution features from the encoder with the feature map output of the bottleneck layer in the decoder. This fusion aids in restoring spatial details that may have been lost during downsampling in the encoder, enabling image reconstruction or segmentation at the original resolution.
By utilizing this amalgamated information, the decoder conducts upsampling to produce a high-resolution output resembling the initial input. Furthermore, following the last decoder block, a 1x1 convolution, indicated by violet arrows in the model's structure, is employed alongside a sigmoid activation. This generates a segmentation mask output sized 256x256x1, containing pixel-wise classifications. This mechanism enables the transmission of information from the contracting path to the expansive path, capturing both feature details and localization. Consequently, it enhances the effectiveness of U-Net for semantic segmentation tasks.

V. RESULTS AND DISCUSSION
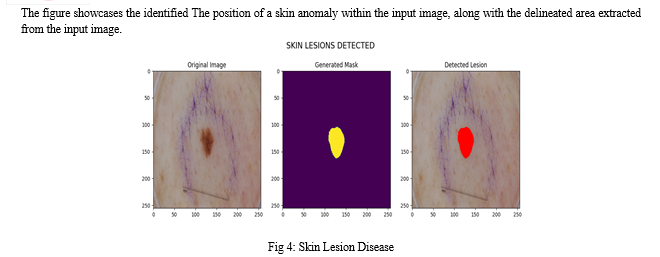
The outcome of our proposed implementation involves selecting between brain tumors or skin lesions. Afterward, we upload the image or input images. From the input image, we receive results indicating the detected tumor or lesion region, along with a masked image. Additionally, we receive information regarding the presence of tumors or lesions in the input image. If no tumor or lesion is detected, we obtain an empty masked image without any segmented regions of tumors or lesions. In such cases, no detected tumor or lesion regions are displayed in the image.
VI. ACKNOWLEDGMENT
We express our gratitude to the researchers and developers behind the U-Net architecture and associated technologies for their invaluable contributions to the advancement of medical image analysis. Furthermore, we extend our sincere appreciation to all participants who contributed to this study, as their cooperation and involvement were indispensable in making this research possible.
Conclusion
Our implementation of the U-Net architecture prioritizes lightweight design without compromising segmentation accuracy, thereby minimizing the need for extensive data augmentation. This framework shows potential for integration into medical settings, where trained physicians could utilize it as a supplementary tool for evaluating patients\' MRI or skin images. While investigations into brain tumour segmentation using deep learning have made considerable strides, additional research is necessary to further advance the field warranted to enhance the network\'s performance, particularly in reducing false negatives and false positives in biomedical image analysis. Future research endeavours could explore the incorporation of disease-specific information for brain tumours or skin lesions, potentially offering substantial benefits to the medical community.
References
[1] Ramin Ranjbarzadeh et al,\"Brain tumor segmentation built upon DL techniques and an attention mechanism using MRI multi-modalities brain images\", Scientific Reports,11 Article number 10930 , May 2021. [2] Lina Liu Ying Y.Tsui et al,\"Skin Segmentation of Lesions Utilizing DL with Auxiliary Task\",J Imaging,April 2021. [3] Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M.,\" Thrun Sophisticated classification of skin cancer akin to that of dermatologists achieved through deep neural networks. Nature\". 2017;542:115–118. Aug 2023, [4] Khosravanian, A., Rahmanimanesh, M., Keshavarzi, P. & Mozaffari, \"Fast level set method for Segmentation of glioma brain tumors utilizing a specific approach superpixel fuzzy clustering and lattice boltzmann method\". Comput. Methods Programs Biomed. 198, 105809,2020. [5] Hihao ,\"Medical Image Segmentation Derived from U-Net\", Journal of Physics:Conference Series,vol.2547 , 2023 [6] Mohammed Khouy ,\"Medical Image Segmentation Using Automatic Optimized U-Net Architecture Based on Genetic Algorithm\",MDPI,Aug 2023. [7] cafer budak,\"Biomedical Image Partitioning Using Modified U-Net\",IIETA,March 2023. [8] Olaf Ronneberger,\"U-Net: Convolutional Networks for Biomedical Imaging Segmentation\",vol.9351,nov 2015.
Copyright
Copyright © 2024 Sailaja Y, Suresh Kumar Gummadi, Harshitha Gorrepati, Deepika Pucha. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET59064
Publish Date : 2024-03-16
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

