Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Computational Insights into the Functional Impact of Missense Mutations in RFX1
Authors: Nidhi S Hallikeri, Kaveri Hallikeri
DOI Link: https://doi.org/10.22214/ijraset.2024.66138
Certificate: View Certificate
Abstract
Genetic variations, particularly non-synonymous SNPs (nsSNPs), can profoundly influence protein function by altering stability, molecular interactions, and regulatory pathways, thereby contributing to disease mechanisms, including cancer. Despite advancements in high-throughput sequencing, the effects of nsSNPs on structure and function, especially in transcriptional regulators like RFX1, remain poorly understood. Addressing these challenges is crucial for uncovering potential biomarkers and therapeutic targets in cancer biology. This study focused on assessing the impact of deleterious nsSNPs in RFX1 on the stability, function, and structure of proteins, with a particular emphasis on its interaction with HDAC1. Using data from the COSMIC database and computational tools such as I-Mutant, MutPred, and Arpeggio, we identified 19 missense mutations predicted to be deleterious, significantly impacting RFX1 function. The most detrimental variants included R495L, V786G, R494C, A715P, Y460C, L812R, R311H, and R311C. Further analysis using STRING highlighted RFX1\'s involvement in gene transcription and chromatin remodeling pathways, mediated through interactions with key regulatory proteins such as CREB1, CIITA, and HDAC1. RFX1 expression analysis revealed its potential as a prognostic biomarker, with statistically significant hazard ratios (HR < 1) observed in breast, ovarian, and pancreatic cancers, suggesting its role as a protective factor in these malignancies. Prognostic evaluations further underscored RFX1\'s potential as a key biomarker in cancer. Future research will focus on experimental validation of these deleterious mutations and expression-based findings to further elucidate their diagnostic and therapeutic relevance.
Introduction
I. INTRODUCTION
The role of histone deacetylases (HDACs) and transcription factors in gene regulation has drawn considerable attention in molecular biology. The RFX family, particularly RFX1, interacts with HDACs to repress gene expression, influencing processes like proliferation, migration, invasion, and metastasis [1], [2], [3]. The interplay between RFX1 and HDACs is critical for normal cellular function and cancer prevention, with RFX1 showing anti-cancer properties such as reduced proliferation, apoptosis induction, and enhanced chemosensitivity [4], [5], [6]. Suppressing HDACs contributes to tumor growth inhibition and immune modulation, further emphasizing the significance of these interactions [7] [8], [9].
RFX1 exhibits therapeutic potential against cancer by targeting pathways involved in cancer progression [4], [10]. Similarly, the inhibition of HDACs has been linked to the inhibition of tumor growth and modulation of the immune response, suggesting the potential of HDAC inhibitors in cancer prevention and treatment [11]. The interaction between RFX1 and HDACs may influence these pathways, making it a crucial focus in the study of cancer prevention and normal cell function. This study analyzed the effects of RFX1 variants to identify deleterious nsSNPs and their impact on protein stability and function. These findings provide a computational basis for further experimental validation, offering insights into RFX1’s role in disease mechanisms.
II. METHODOLOGY
A. Data collection
A total of 243 non-synonymous missense mutations in the RFX1 gene were curated from the COSMIC database. The amino acid sequence of RFX1 (P22670, RFX1_HUMAN) was retrieved from the Swiss-Prot database. Crystal structures of RFX1 (PDB ID: 1DP7) and HDAC1 (PDB IDs: 6Z2J, 6Z2K) were sourced from the RCSB PDB and used for non-covalent interaction analysis and molecular docking. Mutant residues were introduced into the structures using PyMol.
B. Prediction of deleterious mutations
A multi-faceted approach was applied to identify the most deleterious nsSNPs. The tools include SIFT [12], PolyPhen-1 & 2 [13], PhD-SNP [14], SNAP [15], PredictSNP [16], and MAPP [17]. We included MutPred2 [18], which predicts changes in post-translational modification features and structural alterations in the protein of interest.
C. Protein stability and structure analysis
I-Mutant suite assessed stability changes of missense mutations at 25°C and pH 7, predicting free energy changes (ΔΔG) and classifying mutations as stabilizing (ΔΔG > 0) or destabilizing (ΔΔG < 0) in kcal/mol [19], [20], [21]. Project HOPE analyzed the structural and functional impacts of missense mutations using UniProtKB data [22]. The Arpeggio webserver evaluated non-covalent interactions, including van der Waals forces, hydrogen bonds, ionic bonds, and hydrophobic contacts, providing insights into protein stability and structure [23]. DynaMut2 predicted the impact of nsSNPs on protein structural stability by analyzing flexibility changes and intramolecular interactions [24]. The secondary structure of the RFX1 protein was predicted using SOPMA, which provides a detailed analysis of the distribution of alpha helices, beta strands, turns, extended regions, bend regions, and random coils [25].
D. Evolutionary conserved residues analysis
The conservation score of each amino acid residue was assessed using Consurf, which evaluates the significance of residues concerning protein structure and function. Scores range from 1 to 9, with lower scores indicating highly conserved positions and higher scores representing rapidly evolving residues [26].
E. PolymiRTS database 3.0
PolymiRTS was used to analyze SNPs and INDEL variations in the 3' UTR regions of Homo sapiens and mouse mRNAs, focusing on microRNA binding sites. These variations can alter miRNA-mRNA interactions, affecting gene expression. Polymorphisms were classified as Class D (Disruptive) or Class C (Conserved), with Class D causing severe disruptions and Class C having a milder impact. Both types can lead to loss of normal miRNA-mediated repression or abnormal gene control. The RFX1 gene (transcript variant 2, ID: NM_002918) was analyzed [27].
F. Protein-protein interactions
STRING compiles protein-protein interaction data derived from empirical evidence, computational predictions, and curated text mining. This resource offers both experimental and theoretical interaction data for RFX1 [28], [29].
G. Kaplan-Meier plotter analysis
The Kaplan-Meier Plotter, a web-based tool, identifies and validates cancer biomarkers through meta-analysis. Overall survival (OS) was defined as the duration from a specific gene expression alteration to the survival of cancer patients. Statistical significance was set at p < 0.05 [30], [31].
H. Protein-protein docking
Interaction analysis was conducted using HDock and ClusPro. The HDOCK server combines template-based and ab initio docking for protein-protein, protein-RNA, and protein-DNA interactions, utilizing receptor and ligand inputs [32], [33], [34], [35]. ClusPro explores protein-protein interactions and evaluates the structural stability of mutants [36].
III. RESULTS
This study explores the effects of specific mutations in the RFX1 gene and their potential impact on HDAC1 interaction, critical for cellular regulation. Using bioinformatics and computational simulations, we examine the structural and functional changes caused by identified missense mutations. nsSNPs from the COSMIC database were analyzed with various computational tools, suggesting that the most harmful substitutions might contribute to cancer progression by altering protein function, possibly promoting cellular immortality. However, the exact biological mechanisms behind these changes remain unclear.
A. Prediction of functional scores of nsSNPs by in-silico tools
44 deleterious missense mutations were identified, each with a PSIC score of 1.000. Functional predictions using tools like PredictSNP, MAPP, PhD-SNP, PolyPhen-1, PolyPhen-2, SIFT, and SNAP indicated that 43, 25, 36, 38, 44, 44, and 32 mutations, respectively, could disrupt RFX1 function. Of the analyzed mutations, 7.8% (19 out of 243) were deemed deleterious across all seven tools (Table I).
TABLE I
PREDICTION OF FUNCTIONAL SCORES OF NSSNPS BY IN-SILICO TOOLS
|
Mutation |
PolyPhen-2 score |
PredictSNP |
MAPP |
PhD-SNP |
PolyPhen-1 |
PolyPhen-2 |
SIFT |
SNAP |
|
A715P |
1 |
87 |
66 |
88 |
59 |
59 |
43 |
81 |
|
A869T |
1 |
87 |
56 |
86 |
74 |
81 |
79 |
81 |
|
E891K |
1 |
87 |
48 |
88 |
74 |
81 |
79 |
87 |
|
G481D |
1 |
87 |
72 |
88 |
74 |
81 |
79 |
81 |
|
G873C |
1 |
87 |
63 |
88 |
74 |
81 |
79 |
85 |
|
L812R |
1 |
87 |
86 |
88 |
74 |
81 |
79 |
81 |
|
N445K |
1 |
87 |
72 |
88 |
74 |
81 |
79 |
72 |
|
P730L |
1 |
87 |
51 |
88 |
74 |
81 |
79 |
72 |
|
R311C |
1 |
87 |
- |
77 |
74 |
81 |
79 |
72 |
|
R311H |
1 |
87 |
- |
73 |
74 |
81 |
43 |
56 |
|
R433C |
1 |
87 |
86 |
86 |
74 |
81 |
79 |
56 |
|
R494C |
1 |
87 |
77 |
88 |
74 |
81 |
79 |
72 |
|
R495L |
1 |
87 |
63 |
89 |
59 |
81 |
79 |
56 |
|
R862W |
1 |
87 |
62 |
86 |
74 |
81 |
79 |
85 |
|
R867C |
1 |
87 |
56 |
77 |
74 |
81 |
79 |
62 |
|
V786G |
1 |
87 |
56 |
88 |
74 |
68 |
79 |
62 |
|
W639C |
1 |
87 |
91 |
88 |
74 |
81 |
79 |
81 |
|
Y460C |
1 |
87 |
72 |
89 |
74 |
81 |
79 |
85 |
|
Y463H |
1 |
87 |
81 |
88 |
74 |
81 |
79 |
85 |
MutPred analysis predicted the disruption of typical RFX1 functions, indicating alterations in features such as methylation, ubiquitination, phosphorylation, catalytic activity, solvent accessibility, and secondary structure (Tables II and III).
TABLE II
DETAILED MUTPRED RESULTS OF RFX1 VARIANTS
|
Mutation |
Sulfation |
Alterations |
Secondary structure |
Glycosylation |
ADP-ribosylation |
|
A71P |
|
|
|
|
|
|
L812R |
|
Altered coiled coil |
Gain of helix |
|
|
|
R311C |
Loss at Y315 |
Altered ordered interface, altered transmembrane protein, Altered disordered interface |
Gain of loop |
Loss at S313 |
Loss at R311 |
|
R311H |
Gain at Y315 |
Altered ordered interface, Altered transmembrane protein |
Gain of loop |
Loss at S313 |
Loss at R311 |
|
R494C |
|
Altered ordered interface, altered transmembrane protein, Altered disordered interface |
Loss of loop |
|
|
|
R495L |
|
Altered disordered interface, Altered DNA binding |
|
|
|
|
V786G |
|
Altered transmembrane protein |
|
|
|
|
Y460C |
|
Gain of helix |
|
|
|
B. Protein stability and structure analysis
I-Mutant 3.0 Suite predicted stability changes for 9 of the 19 nsSNPs of RFX1 (Table II). These mutations (A869T, E891K, G481D, L812R, N445K, R433C, R862W, R867C, and Y463H) may not significantly affect RFX1 stability but could disrupt its dynamics and interaction with HDAC1.
TABLE III
ANALYSIS OF STABILITY EFFECT OF RFX1 VARIANTS BY I-MUTANT SUITE
|
Mutation |
ΔG (Kcal/mol) |
Effect |
|
p.A715P |
-1.25 |
Destabilizing |
|
p.L812R |
0.2 |
Stabalizing |
|
p.R311C |
-0.28 |
Destabilizing |
|
p.R311H |
-0.93 |
Destabilizing |
|
p.R494C |
-3.26 |
Destabilizing |
|
p.R495L |
-2.39 |
Destabilizing |
|
p.V786G |
-5.11 |
Destabilizing |
|
p.Y460C |
-1.01 |
Destabilizing |
Project HOPE and DynaMut2 assessed structural changes, revealing alterations in size, charge, hydrophobicity, salt-bridges, and hydrogen bonds (Table IV, Fig. I). Most mutations affected hydrogen bond formation and ionic interactions due to changes in size, charge, and hydrophobicity.
TABLE IV
HOPE PREDICTION OF SELECTED RFX1 DELETERIOUS NSSNPS
|
RFX1 Varients |
Structural changes |
Altered properties |
|
A715P
|
|
The mutant residue is larger than the wild-type residue, causing structural bumps. This disrupts the alpha helix, potentially leading to severe effects on the protein's structure. |
|
L812R |
|
The mutant residue is larger and positively charged compared to the neutral wild-type residue. Additionally, the mutant residue is less hydrophobic, leading to the loss of hydrophobic interactions, both in the protein core and on the surface. |
|
R311C
|
|
The mutant residue is smaller and neutral, compared to the positively charged wild-type residue. It is also more hydrophobic, which may disrupt hydrogen bonds and affect proper folding. |
|
R311H
|
|
The mutant residue is smaller and neutral, potentially leading to the loss of interactions. The loss of the wild-type residue's positive charge may also disrupt interactions with other molecules or residues. |
|
R494C
|
|
The mutant residue is smaller, neutral, and more hydrophobic than the wild-type, which may lead to loss of interactions, disrupt hydrogen bonds, and affect proper folding. |
|
R495L
|
|
The mutant residue is smaller, neutral, and more hydrophobic than the wild-type, potentially leading to loss of interactions, disruption of hydrogen bonds, and impaired folding. |
|
V786G
|
|
The smaller mutant residue leads to loss of interactions, while the decreased hydrophobicity results in the loss of hydrophobic interactions both in the core and on the surface. |
|
Y460C |
|
The smaller mutant residue may lead to loss of interactions, while its increased hydrophobicity could disrupt hydrogen bonds and affect proper folding. |
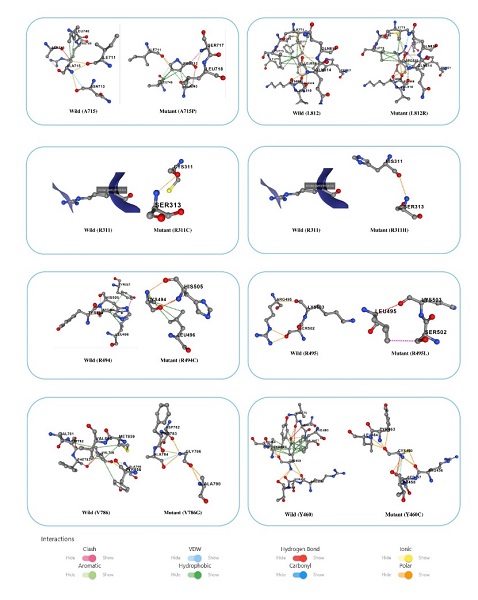
Fig. I Structural comparison of wild-type RFX1 and its variants
The Arpeggio web server categorized the atomic interactions of RFX1 variants, highlighting deviations in hydrogen bonds, hydrophobic interactions, ionic bonds, Van der Waals interactions, polarity, and aromatic contacts that contributed to instability (Table V).
TABLE V
ARPEGGIO PREDICTION OF INTERATOMIC INTERACTIONS OF NATIVE AND MUTANT RFX1
|
Mutations |
Hydrogen bonds |
Hydrophobic contacts |
Van der Waals interactions |
Ionic interactions |
Polar contacts |
Aromatic contacts |
|
RFX1 |
81 |
175 |
102 |
17 |
148 |
299 |
|
A715P |
567 |
1101 |
325 |
64 |
899 |
72 |
|
L812R |
569 |
7090 |
323 |
64 |
901 |
72 |
|
R311C |
569 |
1099 |
323 |
64 |
901 |
72 |
|
R311H |
569 |
1099 |
323 |
64 |
901 |
72 |
|
R494C |
569 |
1098 |
323 |
64 |
901 |
72 |
|
R495L |
568 |
1104 |
323 |
64 |
899 |
72 |
|
V786G |
569 |
1094 |
323 |
64 |
901 |
72 |
|
Y460C |
569 |
1079 |
323 |
64 |
899 |
69 |
C. Analysis of conserved residues of RFX1
The Consurf results predicted that phylogenetically, residues A715, A869, E891, G481, G873, L812, N445, P730, R311, R443, R494, R495, R862, R867, V786, Y460 and Y463, were highly conserved (Fig. II). The highly conserved amino acid residues are not prone to frequent mutational events, and in case of a mutation, it can cause a severe impact on protein structure, function and interaction. In contrast, mutations in less conserved regions may have less significant effects.

Fig. II Conservation analysis of RFX1 using ConSurf
D. Analysis of protein secondary structure
Among 19 highly deleterious nsSNPs, SOPMA (Fig. III) predicted altered secondary structures that lead to 52.6% residues to be in α-helices (A715, E891, G481, G873, L812, R862, V786, W639, Y460, Y463), 31.6% in random coils (A869, N445, P730, R311, R433, R867) and 10.5% extended strands (R494 and R495).

Fig. III Secondary structure prediction of RFX1 using SOPMA
E. Prediction of functional SNPs located in 3′UTRs region
Among all the SNPs in the 3′UTR region of the RFX1 gene, 11 functional SNPs were predicted to affect the miRNA target sites. Seven SNPs, rs148919451, rs1048979, rs115028104, rs10416669, rs116463743, rs61497567 and rs188167241 disrupt miRNA conserved sites (Table VI). Higher conservation scores (9–13) the importance of miRNA binding site and mutations in these regions could lead to functional disruption.
TABLE VI
POLYMIRTS DATABASE PREDICTION RESULTS
|
dbSNP ID |
miR ID |
Conservation |
miRSite |
Function Class |
|
rs148919451 |
hsa-miR-338-5p |
13 |
ctgaccTATTGTA |
D |
|
hsa-miR-370-5p |
7 |
cTGACCTAttgta |
D |
|
|
hsa-miR-6826-5p |
12 |
ctgaCCTATTGta |
D |
|
|
hsa-miR-4506 |
10 |
ctgACCCATTgta |
C |
|
|
hsa-miR-10b-3p |
5 |
AATCTGAcctatt |
D |
|
|
hsa-miR-370-5p |
10 |
aatcTGACCTAtt |
D |
|
|
hsa-miR-6801-5p |
5 |
aaTCTGACCtatt |
D |
|
|
rs1048979 |
hsa-miR-4436b-3p |
10 |
aatCTGCCCTAtt |
C |
|
hsa-miR-4632-5p |
10 |
aatCTGCCCTAtt |
C |
|
|
hsa-miR-4761-3p |
10 |
aatcTGCCCTAtt |
C |
|
|
hsa-miR-6735-5p |
10 |
aatCTGCCCTAtt |
C |
|
|
hsa-miR-6879-5p |
8 |
aatCTGCCCTAtt |
C |
|
|
hsa-miR-7843-5p |
8 |
aatCTGCCCTAtt |
C |
|
|
rs115028104 |
hsa-miR-548as-3p |
9 |
cttgttGGGTTTT |
D |
|
hsa-miR-548at-3p |
9 |
cttgttCGGTTTT |
C |
|
|
hsa-miR-548ay-3p |
9 |
cttgttCGGTTTT |
C |
|
|
rs10416669 |
hsa-miR-4467 |
3 |
gtgagCGCCGCCc |
D |
|
hsa-miR-1178-3p |
5 |
GTGAGCAccgccc |
C |
|
|
hsa-miR-1250-5p |
3 |
gtgAGCACCGccc |
C |
|
|
hsa-miR-3907 |
3 |
gtGAGCACCgccc |
C |
|
|
hsa-miR-4746-3p |
3 |
gtgaGCACCGCcc |
C |
|
|
rs116463743 |
hsa-miR-6715a-3p |
9 |
cgccgcTGGTTTG |
D |
|
hsa-miR-4467 |
3 |
CGCCGCCggtttg |
C |
|
|
rs61497567 |
hsa-miR-4732-3p |
7 |
cctcaGTCAGGGc |
C |
|
hsa-miR-7976 |
11 |
cctcagTCAGGGC |
C |
|
|
rs188167241 |
hsa-miR-6797-5p |
4 |
ccgccaTCCCTCC |
C |
F. Protein-protein interactions analysis
The mutation may change the structure of a protein and thus the function of protein may change. To investigate the interaction of RFX1 with various proteins, the STRING server was used. The interaction analysis revealed that RFX1 is related to Cyclic AMP-responsive element-binding protein 1, Regulatory factor X-associated protein, MHC class II transactivator, DNA-binding protein RFXANK, HDAC1 Histone deacetylase 1, Serine/threonine-protein kinase ATR, Histone-lysine N-methyltransferase SUV39H1, Ribosomal protein L30, Forkhead box protein J1, DNA (cytosine-5)-methyltransferase 1 (Fig. IV).
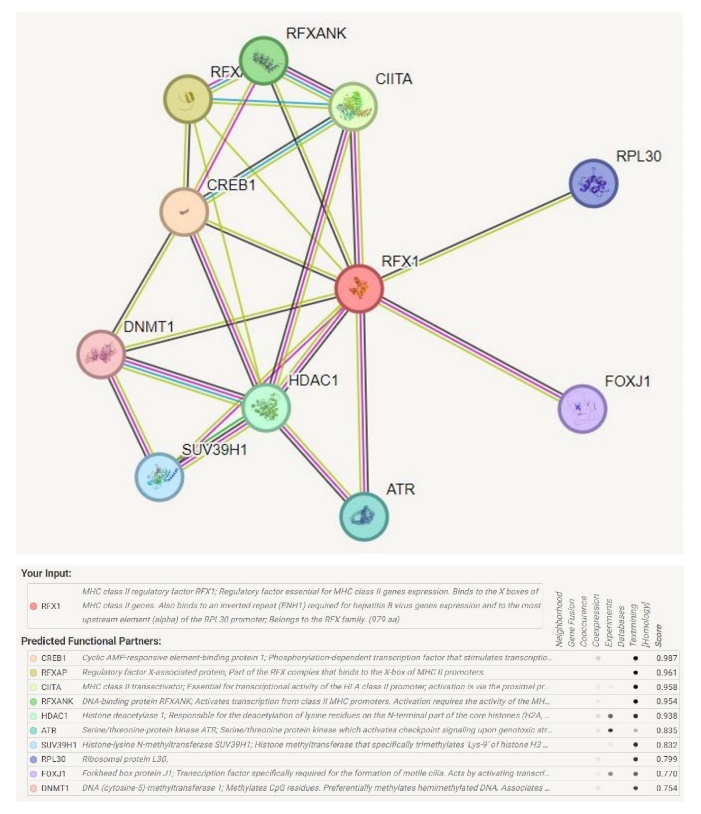
Fig. IV Protein-protein interaction of RFX1 with key regulatory proteins
G. Impact of RFX1 Expression Levels on Overall Survival (OS) in Patients with Various Cancers
In breast cancer, RFX1 expression was associated with a hazard ratio (HR) of 0.76 (95% CI, 0.69-0.85) and a logrank p-value of 0.00000023. Similarly, in ovarian cancer, RFX1 showed a HR of 0.77 (95% CI, 0.67-0.88) and a logrank p-value of 0.00014, also statistically significant. In contrast, for lung cancer, RFX1 had a HR of 1.12 (95% CI, 0.99-1.26) and a logrank p-value of 0.065, suggesting no significant effect. In gastric cancer, RFX1 expression had a HR of 1.65 (95% CI, 1.39-1.95) with a logrank p-value of 0.0000000072, indicating a significant association. For pancreatic cancer, the HR was 0.78 (95% CI, 0.67-0.91) with a p-value of 0.00141, in AML HR was 0.8 (95% CI, 0.69-0.91) with a p-value of 0.001, and in myeloma, the HR was 0.72 (95% CI, 0.57-0.9) with a p-value of 0.0037, all showing indicating a statistically significant relationship between higher RFX1 expression and improved survival rates (Fig. V). The results showed that RFX1 deregulation has distinct implications in different types of cancers. This study shows, the RFX1 deregulation can serve as a prognostic marker for patients with breast, ovarian, gastric, pancreatic, AML and myeloma cancers but not for lung cancer.
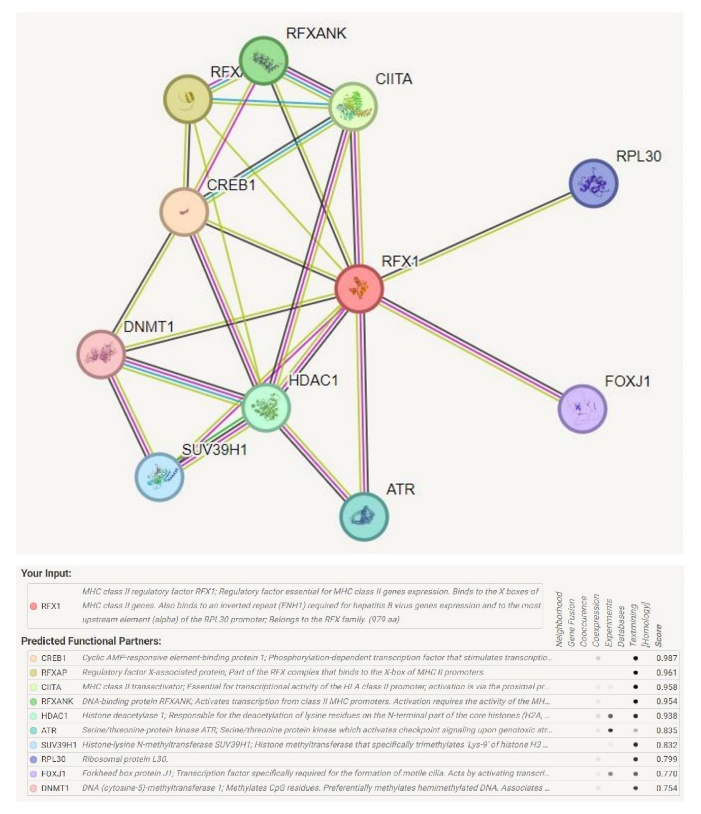
Fig. V Kaplan-Meier survival analysis of OS patients based on RFX1 expression levels
H. Protein-protein docking
Through the present study we try to establish wild type and mutant RFX1-HDAC1 interactions. RFX1 and RFX1 binding domain (PBD) of HDAC1 were docked with HDock and ClusPro as shown in Table VII.
TABLE VII
PROTEIN-PROTEIN INTERACTION BINDING SCORES OF RFX1 WITH RFX1 BINDING DOMAIN OF HDAC1
|
Mutations |
HDock Docking score |
ClusPro Centre |
ClusPro Lowest energy |
|
RFX1 w/HDAC1w |
-248.34 |
-776.0 |
-859.4 |
|
A715P/HDAC1w |
-248.45 |
-853.5 |
-904.1 |
|
L812R/HDAC1w |
-237.58
|
-853.5 |
-904.1 |
|
R311C/HDAC1w |
-255.51 |
-773.4 |
-931.3 |
|
R311H/HDAC1w |
-241.71 |
-752.2 |
-935.0 |
|
R494C/HDAC1w |
-230.77 |
-848.7 |
-899.9 |
|
R495L/HDAC1w |
-236.32 |
-852.4 |
-902.9 |
|
V786G/HDAC1w |
-249.33 |
-853.5 |
-904.1 |
|
Y460C/HDAC1w |
-250.30 |
-853.6 |
-904.1 |
*w-wild/ native conformation.
Conclusion
Identifying nsSNPs is essential for exploring diagnostic markers and pharmacotherapy targets, as these mutations affect expression, stability, folding, interactions, and drug response. This study examines RFX1-HDAC1 interaction changes due to mutations, identifying 19 harmful mutations in RFX1. Mutant Suite analysis of RFX1 variants identified destabilizing (R494C, V786G) and stabilizing mutations (Y463H) that affect protein stability. Destabilizing mutations, particularly p.V786G, may disrupt folding and contribute to disease [37], while stabilizing mutations (Y463H) may alter RFX1’s functional dynamics. MutPred analysis revealed impacts of mutations like G481D, R311C, and R311H on protein stability, DNA binding, and transmembrane domains, affecting RFX1\'s regulatory functions [38], [39]. Structural changes such as loops or helices (G873C, R494C) disrupted conformational flexibility and stability [40], [41]. SNPs like A869T, G481D, and L812R altered PTM sites, affecting protein stability and cellular interactions [42]. Y463H influenced metal binding and transmembrane properties, while G481D and R495L likely impaired transcriptional functions [43]. Project HOPE and DynaMut2 analyses indicated that deleterious nsSNPs affect protein stability and functionality by altering residue size, hydrophobicity, and charge. Mutations such as A715P and A869T disrupted alpha-helical structures, and charge alterations like E891K could disrupt ligand-binding regions [44], [45]. R494C and Y460C caused loss of hydrogen bonds and hydrophobic interactions, affecting folding, stability, and functionality. Arpeggio analysis showed increased hydrogen bonds, polar contacts, and hydrophobic interactions in mutant proteins, suggesting structural compensations [46], [47]. Mutations like A715P, G481D, and L812R displayed elevated hydrogen bonds and polar contacts, whereas L812R showed excessive hydrophobic interactions [48]. R495L and Y463H had reduced aromatic contacts, impairing molecular interactions. SOPMA analysis revealed significant changes in secondary structures, with 52.6% forming ?-helices, 31.6% random coils, and 10.5% extended strands. Increased ?-helices suggest compensatory stabilization, while random coils may compromise stability and DNA-binding affinity [49]. These align with findings from Arpeggio, where ?-helices exhibited increased hydrogen bonding and polar contacts, supporting helical stability and random coils, contribute to reduced rigidity and impaired function. Mutations like L812R, with excessive hydrophobic contacts, could lead to abnormal rigidity, disrupting protein dynamics. Functional analysis of SNPs in the 3? UTR of RFX1 identified 11 variants affecting miRNA target sites. Disruptions in conserved miRNA binding sites and 26 novel miRNA target sites suggest shifts in post-transcriptional regulation, influencing gene expression patterns [50]. 26 novel miRNA target sites were identified, indicating shifts in post-transcriptional regulation that could introduce new regulatory dynamics and influence gene expression patterns. Variants like rs148919451, rs115649319, and rs191735232 disrupted miRNA binding, highlighting the role of non-coding SNPs in gene regulation and disease [51]. STRING analysis of RFX1 reveals connections with regulatory proteins like CREB1, CIITA, and HDAC1, linking it to gene transcription and chromatin remodeling pathways. HDAC1, a histone deacetylase, suggests a regulatory link to chromatin modification, potentially impacting gene silencing or activation states. RFX1 mutations may impact cellular stress responses by interacting with DNA damage response protein ATR and alter gene expression epigenetically. RFX1 expression analysis reveals its potential as a prognostic biomarker. Higher RFX1 expression links to better overall survival (OS) in breast, ovarian, pancreatic cancers, AML, and myeloma (Fig. VII). The inverse relationship in gastric cancer, where high RFX1 expression is associated with poorer outcomes, indicates RFX1 may act differently in cancers with a more inflammatory microenvironment and function of RFX1 differs depending on the cancer type, potentially due to variations in tumor microenvironments or genetic backgrounds that influence RFX1 pathways [4]. Statistically significant hazard ratios in breast, ovarian, and pancreatic cancers indicate RFX1\'s potential as a protective factor. These findings align with prior studies that suggest RFX1 might inhibit tumor progression through pathways affecting cell proliferation or immune response [10]. The lack of a statistically significance in lung cancer implies that RFX1 may not have a universal prognostic value. HDock and ClusPro showed alterations in RFX1-HDAC1 binding, with mutations like R311C and R311H enhancing binding, while R494C and R495L reduced binding stability. In conclusion, RFX1 nsSNPs identified in cancer affect protein structure and function. These findings highlight the impact of highly deleterious mutations on RFX1’s. Future experimental studies, such as site-directed mutagenesis and functional assays, yeast hybridization and post-translational modification (PTM) studies, could further prioritize these SNPs as potential molecular markers for cancer diagnostics.
References
[1] S. Bahl and E. Seto, ‘Regulation of histone deacetylase activities and functions by phosphorylation and its physiological relevance’, Cellular and Molecular Life Sciences, vol. 78, no. 2, pp. 427–445, Jan. 2021, doi: 10.1007/s00018-020-03599-4.
[2] D. Sugiaman-Trapman et al., ‘Characterization of the human RFX transcription factor family by regulatory and target gene analysis’, BMC Genomics, vol. 19, no. 1, p. 181, Dec. 2018, doi: 10.1186/s12864-018-4564-6.
[3] Y. Xu, P. K. Sengupta, E. Seto, and B. D. Smith, ‘Regulatory Factor for X-box Family Proteins Differentially Interact with Histone Deacetylases to Repress Collagen ?2(I) Gene (COL1A2) Expression’, Journal of Biological Chemistry, vol. 281, no. 14, pp. 9260–9270, Apr. 2006, doi: 10.1074/jbc.M511724200.
[4] J. Issac, P. S. Raveendran, and A. V. Das, ‘RFX1: a promising therapeutic arsenal against cancer’, Cancer Cell Int, vol. 21, no. 1, p. 253, Dec. 2021, doi: 10.1186/s12935-021-01952-6.
[5] R. K. Alseksek, W. S. Ramadan, E. Saleh, and R. El-Awady, ‘The Role of HDACs in the Response of Cancer Cells to Cellular Stress and the Potential for Therapeutic Intervention’, Int J Mol Sci, vol. 23, no. 15, p. 8141, Jul. 2022, doi: 10.3390/ijms23158141.
[6] R. Hai et al., ‘The emerging roles of HDACs and their therapeutic implications in cancer’, Eur J Pharmacol, vol. 931, p. 175216, Sep. 2022, doi: 10.1016/j.ejphar.2022.175216.
[7] M.-Q. Shi et al., ‘Advances in targeting histone deacetylase for treatment of solid tumors’, J Hematol Oncol, vol. 17, no. 1, p. 37, May 2024, doi: 10.1186/s13045-024-01551-8.
[8] S. Ropero and M. Esteller, ‘The role of histone deacetylases (HDACs) in human cancer’, Mol Oncol, vol. 1, no. 1, pp. 19–25, Jun. 2007, doi: 10.1016/j.molonc.2007.01.001.
[9] P. Wang, Z. Wang, and J. Liu, ‘Role of HDACs in normal and malignant hematopoiesis’, Mol Cancer, vol. 19, no. 1, p. 5, Dec. 2020, doi: 10.1186/s12943-019-1127-7.
[10] Z. Cui et al., ‘Pan-cancer investigation of RFX family and associated genes identifies RFX8 as a therapeutic target in leukemia’, Heliyon, vol. 10, no. 15, p. e35368, Aug. 2024, doi: 10.1016/j.heliyon.2024.e35368.
[11] G. Shanmugam, S. Rakshit, and K. Sarkar, ‘HDAC inhibitors: Targets for tumor therapy, immune modulation and lung diseases’, Transl Oncol, vol. 16, p. 101312, Feb. 2022, doi: 10.1016/j.tranon.2021.101312.
[12] E. Capriotti, R. B. Altman, and Y. Bromberg, ‘Collective judgment predicts disease-associated single nucleotide variants’, BMC Genomics, vol. 14, no. Suppl 3, p. S2, 2013, doi: 10.1186/1471-2164-14-S3-S2.
[13] I. A. Adzhubei et al., ‘A method and server for predicting damaging missense mutations’, Nat Methods, vol. 7, no. 4, pp. 248–249, Apr. 2010, doi: 10.1038/nmeth0410-248.
[14] R. Calabrese, E. Capriotti, P. Fariselli, P. L. Martelli, and R. Casadio, ‘Functional annotations improve the predictive score of human disease-related mutations in proteins’, Hum Mutat, vol. 30, no. 8, pp. 1237–1244, Aug. 2009, doi: 10.1002/humu.21047.
[15] P. C. Ng, ‘SIFT: predicting amino acid changes that affect protein function’, Nucleic Acids Res, vol. 31, no. 13, pp. 3812–3814, Jul. 2003, doi: 10.1093/nar/gkg509.
[16] J. Bendl et al., ‘PredictSNP: Robust and Accurate Consensus Classifier for Prediction of Disease-Related Mutations’, PLoS Comput Biol, vol. 10, no. 1, p. e1003440, Jan. 2014, doi: 10.1371/journal.pcbi.1003440.
[17] E. A. Stone and A. Sidow, ‘Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity’, Genome Res, vol. 15, no. 7, pp. 978–986, Jul. 2005, doi: 10.1101/gr.3804205.
[18] B. Li et al., ‘Automated inference of molecular mechanisms of disease from amino acid substitutions’, Bioinformatics, vol. 25, no. 21, pp. 2744–2750, Nov. 2009, doi: 10.1093/bioinformatics/btp528.
[19] C. Savojardo, P. Fariselli, and R. Casadio, ‘Improving the detection of transmembrane ?-barrel chains with N-to-1 extreme learning machines’, Bioinformatics, vol. 27, no. 22, pp. 3123–3128, Nov. 2011, doi: 10.1093/bioinformatics/btr549.
[20] P. Fariselli, C. Savojardo, P. L. Martelli, and R. Casadio, ‘Grammatical-Restrained Hidden Conditional Random Fields for Bioinformatics applications’, Algorithms for Molecular Biology, vol. 4, no. 1, p. 13, Dec. 2009, doi: 10.1186/1748-7188-4-13.
[21] C. Savojardo, P. Fariselli, and R. Casadio, ‘BETAWARE: a machine-learning tool to detect and predict transmembrane beta-barrel proteins in prokaryotes’, Bioinformatics, vol. 29, no. 4, pp. 504–505, Feb. 2013, doi: 10.1093/bioinformatics/bts728.
[22] H. Venselaar, T. A. te Beek, R. K. Kuipers, M. L. Hekkelman, and G. Vriend, ‘Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces’, BMC Bioinformatics, vol. 11, no. 1, p. 548, Dec. 2010, doi: 10.1186/1471-2105-11-548.
[23] H. C. Jubb, A. P. Higueruelo, B. Ochoa-Montaño, W. R. Pitt, D. B. Ascher, and T. L. Blundell, ‘Arpeggio: A Web Server for Calculating and Visualising Interatomic Interactions in Protein Structures’, J Mol Biol, vol. 429, no. 3, pp. 365–371, Feb. 2017, doi: 10.1016/j.jmb.2016.12.004.
[24] C. H. M. Rodrigues, D. E. V. Pires, and D. B. Ascher, ‘
Copyright
Copyright © 2025 Nidhi S Hallikeri, Kaveri Hallikeri. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET66138
Publish Date : 2024-12-26
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online









