Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
COVID-19 and Other Pneumonia Diagnosis Using CNN
Authors: J Rakesh
DOI Link: https://doi.org/10.22214/ijraset.2022.47249
Certificate: View Certificate
Abstract
In the present scenario, a novel virus called COVID-19 is one of the substantial snags to the humans in this contemporary era. The present pace at which the virus is spreading has led to a significant spike in death rates across the globe, including in all countries and towns. In order to treat the ailment, one must therefore come up with a quick and accurate diagnosis. Deep learning has made a significant contribution to medical research, opening up new paths and opportunities for disease diagnosis methods. To counteract the COVID-19 outbreak, here we create a classifier distinguishing between positive and negative COVID scans. This study suggests a technique that utilizes chest X-ray images to first classify a patient into one of the bacterial or viral classifications, then further subclassifies them into one of the COVID-19 classes (Positive-1 & Negative-0).
Introduction
I. INTRODUCTION
An infectious sickness known as the new coronavirus (COVID-19) started in late December 2019 and has since expanded worldwide throughout the globe. The WHO declared this novel virus know as COVID-19 to be a pandemic on 11th March, 2020. This illness continues to have a devastating impact on the health and well-being of individuals worldwide. Creating a reliable classification system is a crucial phase in the COVID-19 battle cycle so that patients can start receiving immediate medical attention, treatment, and control transmission.
During this little time frame, scientists and researchers have created a variety of diagnostic methods and categorization methods. For instance, one of the most widely used method that is RT-PCR is a critical is an essential screening technique screening tool to identify SARS-COV-2 and as well as COVID-19. Despite being the most used screening approach for the identification of COVID-19, the RT-PCR assay has certain limitations.
There is a lot of time and effort involved in the RT-PCR process, and it may be extremely difficult so in order to identify COVID-19, medical experts have turned to chest radiography imaging techniques including CT or chest x-ray visuals images. In the case of COVID-19, for instance, CT chest scans were found to be more effective than RT-PCR in making a diagnosis. Researchers found that chest CT has a higher sensitivity for identifying COVID-19.
RT-PCR genetic diagnosis are presently the most commonly used to diagnose COVID-19. These evaluations are highly precise. It is possible to identify and test viruses in patient samples, even if they are very little. It is important to remember that now the PCR analysis is quite challenging, requires a lot of time, and is very costly. As a direct consequence of this, not all medical institutions have the necessary equipment to execute it out. In light of these limitations, radiographic scanning may serve as a stand-in diagnostic approach for the sickness. It is possible to evaluate chest radiography images in order to search for the new coronavirus or its symptoms. According to studies, this family of viruses exhibits a strong radiographic manifestation [4,5,6,7,8,9,10,11].
As a result, it may be argued that categorization using radiographic images, such as a chest X-ray (CXR), is not only significantly quicker and less expensive than a PCR test, but also more accurate. Additionally, chest X-rays may be obtained in practically every clinic and are more affordable than other radiological examinations like CT scans.
The only issue with CXR-based detection of COVID-19 patients, particularly in remote places, is that qualified physicians might not always be accessible. Additionally, many professionals lack prior expertise with COVID-19 positive patient CXRs and are unfamiliar with the radiological symptoms associated with COVID-19. Therefore, utilising CXR pictures, we have suggested a straightforward and low-cost deep learning-based method to categorise COVID-19 +ve and -ve cases. This method allows for the quick and nearly exact identification of COVID-19 positive patients.
As a part of this research, We have also supplied a tool to this study that may be used to identify COVID-19 positive patients. This deep learning-based tool will always provide an opinion without the need for human interaction, regardless of the presence or absence of a radiologist or any discrepancies between the doctors' opinions.
II. STATE OF THE ART (LITERATURE SURVEY)
A. Purpose
In light of this, research that were listed in trustworthy databases were retrieved, examined, judged, and synthesised. Comparing the sensitivities, specificities, and AUC Scores of the final 11 articles that were shortlisted. Their models were evaluated, and the best two were selected to form the ensemble for the optimal strategy.
B. Results Obtained
The results obtained is shown below :
TABLE I. Comparing Various DL Model’s
|
Dataset |
Deep Learning Model |
Sensiti-vity |
Specifi-city |
AUC |
|
COVID-19 / NON-COVID PNEUMONIA/NORMAL |
MICROSOFT CUSTOM VISION |
100 |
95 |
- |
|
COVID-19 / NON-COVID PNEUMONIA/NORMAL |
Attention - based VGG-16 |
99.3 |
99.98 |
- |
|
COVID-19 / NORMAL |
ResNet50 |
92.4 |
92.4 |
0.94 |
|
COVID-19 / VIRAL-PNEUMONIA / NORMAL |
VGG-16 |
- |
- |
0.9978 |
|
COVID-19 / VIRAL / BACTERIAL -PNEUMONIA / NORMAL |
DenseNet-201 |
- |
- |
- |
|
COVID-19 / NON-COVID |
HRNet |
98.52 |
98.52 |
- |
|
COVID/BACTERIAL PNEUMONIA / VIRAL PNEUMONIA / NORMAL |
VGG-16 |
93 |
93 |
0.85 |
|
COVID-19 / NON-COVID |
Faster R-CNN |
95.65 |
95.48 |
- |
|
COVID/BACTERIAL PNEUMONIA / VIRAL PNEUMONIA / NORMAL |
IKONOS |
97.7 |
99.3 |
- |
|
COVID-19 / PNEUMONIA / NORMAL |
DarkNet-19 |
95.13 |
95.3 |
- |
|
COVID-19 / NON-COVID |
Residual Att Net |
100 |
96 |
1 |
C. AI vs Radiologist Perfomance
TABLE I. AI Vs Radiologist
|
AI PERFOMANCE |
RADIOLOGIST PERFOMANCE |
||
|
SENSITIVITY |
SPECIFICITY |
SENSITIVITY |
SPECIFICITY |
|
0.92 |
0.97 |
0.90 |
0.88 |
|
0.92 |
0.99 |
0.77 |
0.90 |
|
0.92 |
0.91 |
0.96 |
0.72 |
|
0.84 |
0.83 |
0.75 |
0.94 |
III. PROPOSED WORK
The objective is to build a classifier that can distinguish between a Healthy and Infected (Viral & Bacterial) Chest X-Ray. Additionally, another model takes in viral chest X-Rays and further divide into one of the COVID-19 classes (Positive & Negative).
At this moment, the general public is only authorized to see a tiny subset of the X-ray pictures associated with the COVID-19 investigation. As a consequence, it is unable to construct these models from the very base. In this investigation, we have used a two-pronged strategy to address the dearth of COVID-19 visual images. Both of these strategies will be addressed in further detail below:
- We construct a slightly different version of the COVID-19 imagery by increasing the total number of samples and improving the data in a variety of ways (such instance by inverting the data, rotating it slightly, or gently distorting it).
- The process of training the model now requires less labelled examples of each category than it did before we performed some fine-tuning on the very last layer of the model. As a direct result of this, we were able to go through the steps more quickly.
A. Block Diagram

B. Dataset
For the initial model which divides the given X-Ray into Healthy, Bacterial or Viral, we use the dataset available on Kaggle.
The data are split into two folders:
- 4672 train photographs are in the folder train_images, of which:
- 1227 images belong to class 0
- 2238 images belong to class 1
- 1207 images belong to class 2
- 1168 train photographs are in the folder test_images
- The class labels for the photos are listed in pairs (file_name, class_id) in the file labels_train.csv.
- Where,
- class_id = 0 if the picture depicts a person who is healthy (normal)
- class_id = 1 if the patient in the picture has bacterial pneumonia
- class_id = 2 if the patient in the picture has viral pneumonia
For second model, which further divides into one of the COVID-19 classes (Positive & Negative). We use another dataset from Kaggle.
5. Folder train_images contains 15264 train images.
6. Folder test_images contains 400 test images.
7. The file train_labels.csv is a CSV file with two columns. The first column lists the names of the photos in the train set, and the second lists their labels. There are two possible labels: 1 and 0. For positive samples, the number is 1, and for negative samples, 0. You must handle uneven data in this challenge because the dataset is unbalanced and almost only 10% of the samples are positive.
C. Deep Learning Model to be used – VGG16
The Visual Geometry Group is referred to as VGG. It is an example of a conventional DCNN architect and consists of numerous layers. The number of convolutional layers determines how "deep" a network is, hence the “VGG-16” and “VGG-19” each contain 16 or 19 layers. Innovative item recognition models are developed with the help of the VGG algorithm. The VGGNet, which was designed as a DNN, outperforms benchmarks in a variety of activities and datasets that are not related to ImageNet. It has remained the most often used paradigm of image recognition.

D. Ensemble Method
- For better accuracy on predictions made the confidence predicted by the model can be ensembled for even more closer/better prediction.
- For this case we use a simple averaging, where the output accuracy of each model is averaged with another to predict the final accuracy.
IV. IMPLEMENTATION
This complete project can be divided into 3 main modules as described below:
- MODULE – I: Build Model for Pneumonia Classification into Healthy, Viral and Bacterial.
- MODULE – II: Build second model for COVID-19 detection.
- MODULE – III: Build a user-friendly dashboard & Integrate the Models into it.
A. Module – I
Begin with data Augmentation, some samples of augmentation are shown below.

V. RESULTS DISCUSSION
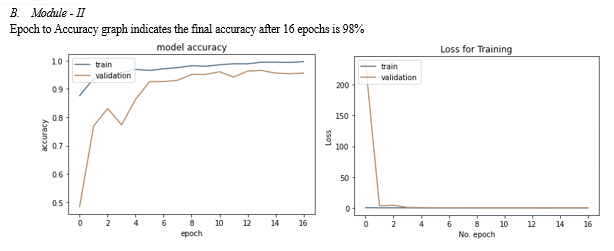
The COVID-19 has recently presented new difficulties in all human beings' daily lives. There are now much more infectious individuals carrying this virus. Therefore, prompt diagnosis and treatment of this infection are quite essential. Within the scope of this investigation, we propose a technique for the prompt and reliable identification of pneumonia. Chest X-rays are used in this procedure for the purpose of classifying patients between two to four distinct groups, with the groups' membership being determined by a variety of different factors. The accuracy of the suggested method is at least 90% and consistently beats the performance of the most advanced approaches currently available for detecting pneumonia.
Conclusion
For the future scope, Additionally, a model can be developed which can predict and work with the GGO (Ground Glass Opacity) along with the pneumonia detected which can help in a better accuracy of the model.
References
[1] Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [CrossRef] [2] Zhong, N.S.; Zheng, B.J.; Li, Y.M.; Poon, L.L.M.; Xie, Z.H.; Chan, K.H.; Li, P.H.; Tan, S.Y.; Chang, Q.; Xie, J.P.; et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 2003, 362, 1353–1358. [CrossRef] [3] Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [CrossRef] [4] WorldOMeter. Coronavirus Live Statistics. 2020. Available online: https://www.worldometers.info/coronavirus/ (accessed on 2 December 2020). [5] Kucirka, L.M.; Lauer, S.A.; Laeyendecker, O.; Boon, D.; Lessler, J. Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction-Based SARS-CoV-2 Tests by Time Since Exposure. Ann. Intern. Med. 2020, 173, 262–267. [CrossRef] [6] Mahase, E. COVID-19: Innova lateral flow test is not fit for “test and release” strategy, say experts. BMJ 2020, 371, m4469. [CrossRef] [7] Zokaeinikoo, M.; Kazemian, P.; Mitra, P.; Kumara, S. AIDCOV: An Interpretable Artificial Intelligence Model for Detection of COVID-19 from Chest Radiography Images. ACM Trans. Manag. Inf. Syst. 2021, 12, 1–20. [CrossRef] [8] Ahmed, S.; Hossain, T.; Hoque, O.B.; Sarker, S.; Rahman, S.; Shah, F.M. Automated COVID-19 Detection from Chest X-Ray Images: A High Resolution Network (HRNet)Approach. SN Comput. Sci. 2021, 2, 1–17. [CrossRef] [9] Yousefzadeh, M.; Esfahanian, P.; Movahed, S.M.S.; Gorgin, S.; Lashgari, R.; Rahmati, D.; Kiani, A.; Nadji, S.A.; Haseli, S.; Hoseinyazdi, M.; et al. Ai-corona: Radiologist-Assistant Deep Learning Framework for COVID-19 Diagnosis in Chest CT Scans. PLoS ONE 2021, 16, e0250952. [10] Shibly, K.H.; Dey, S.K.; Islam, M.T.U.; Rahman, M.M. COVID Faster R-CNN: A Novel Framework to Diagnose Novel Coronavirus Disease (COVID-19) in X-Ray Images. Inform. Med. Unlocked 2020, 20, 100405. [CrossRef] [11] Pesapane, F.; Codari, M.; Sardanelli, F. Artificial intelligence in medical imaging: Threat or opportunity? Radiologists again at the forefront of innovation in medicine. Eur. Radiol. Exp. 2018, 2, 35. [CrossRef] [12] Duda, R.O.; Hart, P.E.; Education, I.C. Machine Learning Artificial Intelligence, Data Scienc. In Pattern Classification and Scene Analysis; Wiley: New York, NY, USA, 1973; Volume 3. [13] Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [CrossRef] Diagnostics 2022, 12, 869 22 of 22 [14] Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G.M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD Statement. BMC Med. 2015, 13, 1. [CrossRef] [15] Nagendran, M.; Chen, Y.; Lovejoy, C.A.; Gordon, A.C.; Komorowski, M.; Harvey, H.; Topol, E.J.; Ioannidis, J.P.A.; Collins, G.S.; Maruthappu, M. Artificial intelligence versus clinicians: Systematic review of design, reporting standards, and claims of deep learning studies. BMJ 2020, 368, m689. [CrossRef] [16] Krarup, M.M.K.; Krokos, G.; Subesinghe, M.; Nair, A.; Fischer, B.M. Artificial Intelligence for the Characterization of Pulmonary Nodules, Lung Tumors and Mediastinal Nodes on PET/CT. Semin. Nucl. Med. 2021, 51, 143–156. [CrossRef] [17] Borkowski, A.A.; Viswanadham, N.A.; Thomas, L.B.; Guzman, R.D.; Deland, L.A.; Mastorides, S.M. Using Artificial Intelligence for COVID-19 Chest X-ray Diagnosis. Fed. Pract. 2020, 37, 398–404. [CrossRef] [18] Keidar, D.; Yaron, D.; Goldstein, E.; Shachar, Y.; Blass, A.; Charbinsky, L.; Aharony, I.; Lifshitz, L.; Lumelsky, D.; Neeman, Z.; et al. COVID-19 Classification of X-ray Images Using Deep Neural Networks. Eur. Radiol. 2021, 31, 9654–9663. [CrossRef] [19] Li, A.C.; Lee, D.T.; Misquitta, K.K.; Uno, K.; Wald, S. COVID-19 Detection from Chest Radiographs Using Machine Learning and Convolutional Neural Networks. medRxiv 2020. [CrossRef] [20] Taresh, M.M.; Zhu, N.; Ali, T.A.A.; Hameed, A.S.; Mutar, M.L. Transfer learning to detect COVID-19 automatically from X-ray images, using convolutional neural networks. Int. J. Biomed. Imaging 2021, 2021. [CrossRef] [21] Kikkisetti, S.; Zhu, J.; Shen, B.; Li, H.; Duong, T. Deep-learning convolutional neural networks with transfer learning accurately classify COVID19 lung infection on portable chest radiographs. PeerJ 2020, 8, e10309. [CrossRef] [22] Gomes, J.C.; Barbosa, V.A.d.F.; de Santana, M.A.; Bandeira, J.; Valenca, M.J.S.; de Souza, R.E.; Ismael, A.M.; dos Santos, W.P. IKONOS: An intelligent tool to support diagnosis of COVID-19 by texture analysis of x-ray images. Res. Biomed. Eng. 2020, 1–14. [23] Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [CrossRef] [24] Sharma, V.; Dyreson, C. COVID-19 Screening Using Residual Attention Network an Artificial Intelligence Approach. arXiv 2020, arXiv:2006.16106. Available online: https://ui.adsabs.harvard.edu/abs/2020arXiv200616106S (accessed on 1 June 2020). [25] Warman, A.; Warman, P.; Sharma, A.; Parikh, P.; Warman, R.; Viswanadhan, N.; Chen, L.; Mohapatra, S.; Mohapatra, S.S.; Sapiro, G. Interpretable Artificial Intelligence for COVID-19 Diagnosis from Chest CT Reveals Specificity of Ground-Glass Opacities. medRxiv 2020. [CrossRef] [26] He, X.; Wang, S.; Shi, S.; Chu, X.; Tang, J.; Liu, X.; Yan, C.; Zhang, J.; Ding, G. Benchmarking Deep Learning Models and Automated Model Design for COVID-19 Detection with Chest CT Scans. medRxiv 2020. [CrossRef] [27] Goel, C.; Kumar, A.; Dubey, S.K.; Srivastava, V. Efficient Deep Network Architecture for COVID-19 Detection Using Computed Tomography Images. medRxiv 2020. [CrossRef] [28] Zheng, C.; Deng, X.; Fu, Q.; Zhou, Q.; Feng, J.; Ma, H.; Liu, W.; Wang, X. Deep Learning-based Detection for COVID-19 from Chest CT using Weak Label. medRxiv 2020. [CrossRef] [29] Lokwani, R.; Gaikwad, A.; Kulkarni, V.; Pant, A.; Kharat, A. Automated Detection of COVID-19 from CT Scans Using Convolutional Neural Networks. arXiv 2020, arXiv:2006.13212. [30] Liu, B.; Liu, P.; Dai, L.; Yang, Y.; Xie, P.; Tan, Y.; Du, J.; Shan, W.; Zhao, C.; Zhong, Q. Assisting Scalable Diagnosis Automatically via CT Images in the Combat against COVID-19. Sci. Rep. 2021, 11, 1–8. [CrossRef] [31] Guiot, J.; Vaidyanathan, A.; Deprez, L.; Zerka, F.; Danthine, D.; Frix, A.-N.; Thys, M.; Henket, M.; Canivet, G.; Mathieu, S.; et al. Development and validation of an automated radiomic CT signature for detecting COVID-19. Diagnostics 2020, 11, 41. [CrossRef] [32] Ko, H.; Chung, H.; Kang, W.S.; Kim, K.W.; Shin, Y.; Kang, S.J.; Lee, J.H.; Kim, Y.J.; Kim, N.Y.; Jung, H.; et al. COVID-19 Pneumonia Diagnosis Using a Simple 2D Deep Learning Framework with a Single Chest CT Image: Model Development and Validation. J. Med. Internet Res. 2020, 22, e19569. [CrossRef]
Copyright
Copyright © 2022 J Rakesh. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET47249
Publish Date : 2022-10-31
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

