Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Deep Learning Driven Liver Tumor Detection and Classification
Authors: Ajjuguttu Triveni Reddy, Dova Bhavana, Athota Venkataramaiah, Ala Gopal, Mrs. B. Lalitha Rajeswari
DOI Link: https://doi.org/10.22214/ijraset.2024.59142
Certificate: View Certificate
Abstract
Liver tumors represent a significant health concern globally, ranking as the fifth most prevalent type of tumor in men and the ninth in women according to recent Global Cancer Statistics for 2018. The current diagnostic methods involve utilizing imaging tests such as Computed Tomography (CT), Magnetic Resonance Imaging (MRI), and ultrasound, often requiring invasive tissue sampling for confirmation. However, these approaches are not only costly but also time-consuming, posing challenges in timely diagnosis and treatment. This paper proposes an innovative solution leveraging deep learning techniques in image processing for liver tumor diagnosis. Specifically, we introduce a Classification Ensemble model combining ResNet, UNet, ResUNet, Xception, and DenseNet architectures. These models are adept at extracting intricate features from medical images, enhancing the accuracy of tumor classification. Additionally, we integrate state-of-the-art object detection models, including FasterRCNN, YOLOv3, and YOLOv5, to enable precise localization and delineation of liver tumors within the images. We focus on the liver tumors such as cholangiocarcinoma and metastatis. Overall, our proposed framework holds promise in revolutionizing liver tumor diagnosis, potentially leading to earlier detection, improved patient outcomes, and reduced healthcare costs.
Introduction
I. INTRODUCTION
The liver, situated beneath the ribcage on the right side of the abdomen, is the body's largest organ. Liver tumors, resulting from abnormal cell growth, are indicative of various liver conditions, including cirrhosis, hepatitis, and liver cancer, which have led to significant mortality rates globally. Comprising the right and left lobes, the liver plays essential roles in bile production for digestion and detoxification processes. Liver metastasis occurs when cancer spreads to or from the liver, commonly originating from colorectal, lung, breast, pancreatic, stomach, esophageal, and melanoma cancers. Cholangiocarcinoma, the second most prevalent primary hepatobiliary tumor, arises from the bile duct and can be intrahepatic or extrahepatic. This paper focuses on metastatic and cholangiocarcinoma cancers, introducing a Classification Ensemble model integrating robust architectures for precise tumor classification. It also incorporates advanced object detection models to accurately localize liver tumors within images. By leveraging deep learning and ensemble methods, the proposed approach aims to streamline the diagnostic process, offering a cost-effective alternative to traditional methodologies. This review emphasizes recent advancements in computer-aided diagnosis of liver cancer, particularly focusing on deep learning-based techniques for liver tumor segmentation in CT images, and outlines future directions in this rapidly evolving field.
II. LITERATURE SURVEY
Liver cancer represents a significant global health concern, with rising incidence rates observed worldwide [3, 9]. Consequently, there is a growing interest in developing precise and effective computer-aided diagnosis (CAD) systems for detecting and segmenting liver tumors on medical imaging, particularly computed tomography (CT) scans. Deep learning techniques, notably convolutional neural networks (CNNs), have emerged as promising tools in this field due to their capability to automatically learn hierarchical features from medical images.
Several studies have concentrated on utilizing CNNs for liver tumor detection and segmentation. Arakeri [1] explores recent advances and future prospects in CAD systems for liver cancer diagnosis on CT images, highlighting the potential of deep learning methods. Similarly, Yasaka et al. [4] conducted a preliminary study using CNNs to differentiate liver masses on dynamic contrast-enhanced CT scans, exhibiting promising results in automated tumor classification.
Segmentation, the process of outlining tumor boundaries, is crucial for precise diagnosis and treatment planning. Numerous approaches have been proposed for liver tumor segmentation using CNNs. For example, Kaluva et al. [2] proposed a 2D-densely connected CNN for automatic liver and tumor segmentation, achieving high accuracy in delineating tumor regions.
Similarly, Todoroki et al. [6] and Li et al. [17] employed deep CNNs for liver tumor candidate detection and segmentation on CT images, underlining the potential of these methods in clinical practice.
Furthermore, researchers have investigated the integration of advanced techniques to enhance the performance of CNN-based segmentation models. Frid et al. [13] explored the use of generative adversarial networks (GANs) for synthetic medical image augmentation, with the aim of improving CNN performance in liver lesion classification tasks. Additionally, Almotairi et al. [22] proposed a modified signet architecture for liver tumor segmentation in CT scans, demonstrating the effectiveness of tailored network architectures in medical imaging tasks.
III. METHODOLOGY
The project consists of several key modules, each with a specific role. Initially, the "Data Exploration" module loads and analyzes data. The "Image Data Generator" preprocesses images for augmentation. Object detection is facilitated through Torchvision. Data is split into train and test sets for model evaluation. For classification, an ensemble of ResNet, UNet, ResUNet, Xception, and DenseNet is employed. For detection, FasterRCNN with ResNet FPN, YOLOv3, and YOLOv5 are used. User interaction is managed by the "User Signup & Login" module, while "User Input" allows users to input data for predictions. The "Classification and Detection " module displays final predictions. These modules collectively offer a comprehensive solution for image tasks. The subsequent sections elaborate on the dataset particulars, preprocessing steps, data augmentation techniques, and the proposed model.
A. Dataset
Experiments are conducted to analyze the robustness of the proposed models, aimed at classifying different types of liver Tumors from CT scan images. Utilizing the liver tumor dataset collected from kaggle consists of 377 CT scan images. Out of these, 144 images belongs to Cholangiocarcinoma and remaining are belongs metastatis carcinoma.
After Data Augumentation, we take 70% of data as training data and 30% as testing data, with the class-wise distribution.This dataset was suitable for the implementation of the model.
B. Preprocessing
Preprocessing plays a crucial role in preparing medical images for accurate diagnosis using a classification ensemble model and object detection algorithms. Initially, the images undergo standardization to ensure consistency across various modalities. This involves normalization to adjust pixel intensities, followed by resizing to a uniform resolution, mitigating discrepancies in image dimensions. To facilitate feature extraction, image augmentation techniques such as rotation, flipping, and scaling are applied to increase the diversity of the training dataset, promoting generalization and robustness of the model. Additionally, anatomical landmarks or region of interest (ROI) annotations are delineated to guide the object detection models in localizing liver tumors accurately.
C. Data Augumentation
The ImageDataGenerator applies several augmentation techniques to input images. These include rescaling, which ensures that pixel values are within a specific range for better numerical stability and convergence during training. Additionally, shearing involves shifting parts of the image along a specified axis, while zooming alters the scale of the image by zooming in or out. Furthermore, horizontal flipping is applied, which mirrors the image horizontally, effectively doubling the dataset size and introducing variability. Ultimately, the class labels undergo label encoding, representing Cholangiocarcinoma and metastatis as 0 and 1 , respectively.
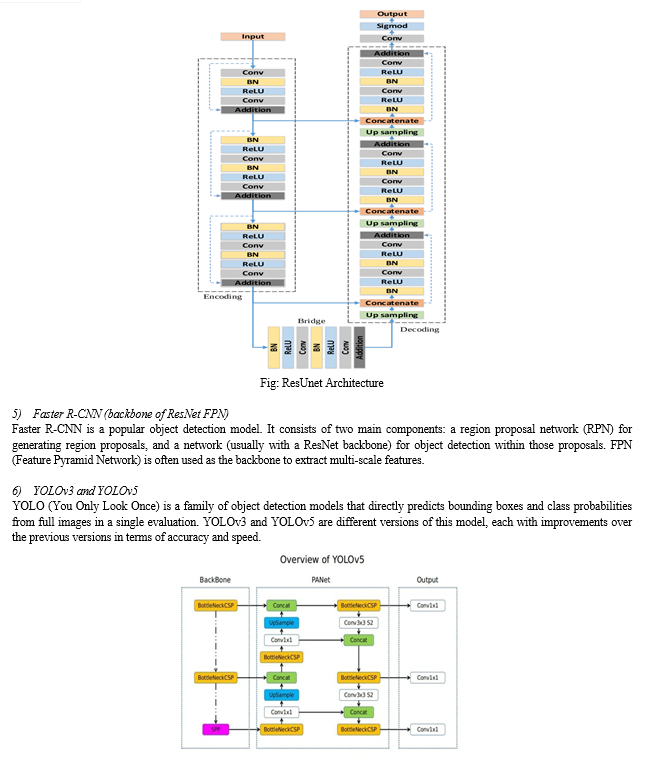
D. Proposed Architecture
This section discusses our proposed system utilizes a Classification Ensemble model incorporating ResNet, UNet, ResUNet, Xception, and DenseNet, alongside object detection models like FasterRCNN, FPN, YOLOv3, and YOLOv5. This fusion enhances accuracy in liver tumor diagnosis by extracting intricate features and precisely localizing tumors within medical images.


2) Modules
- Data exploration: using this module we will load data into system
- Image Data Generator for re-scaling, shear, zoom, flip, reshape.
- Torchvision for detection.
- Splitting data into train & test: using this module data will be divided into train & test
- Model generation: Model building
- Classification- Ensemble of ResNet + UNet – ResUNet, Xception, DenseNet
- Detection - FasterRCNN backbone of ResNet FPN, YoloV3, YoloV5
- User signup & login: Using this module will get registration and login
- User input: Using this module will give input for prediction
- Prediction: final predicted displayed
During the training phase, our system utilizes a varied dataset containing liver tumor images acquired from different imaging modalities. To ensure consistency across models, we preprocess the dataset for uniformity. Each model, including ResNet, UNet, ResUNet, Xception, DenseNet, FasterRCNN, FPN, YOLOv3, and YOLOv5, undergoes training individually using its specific architecture. We employ transfer learning by initializing the models with pre-trained weights from datasets like ImageNet to expedite convergence. Following separate training sessions, we merge the predictions of all models using an ensemble approach. We use techniques like majority voting or weighted averaging to combine outputs, leveraging each model's strengths while mitigating its weaknesses.
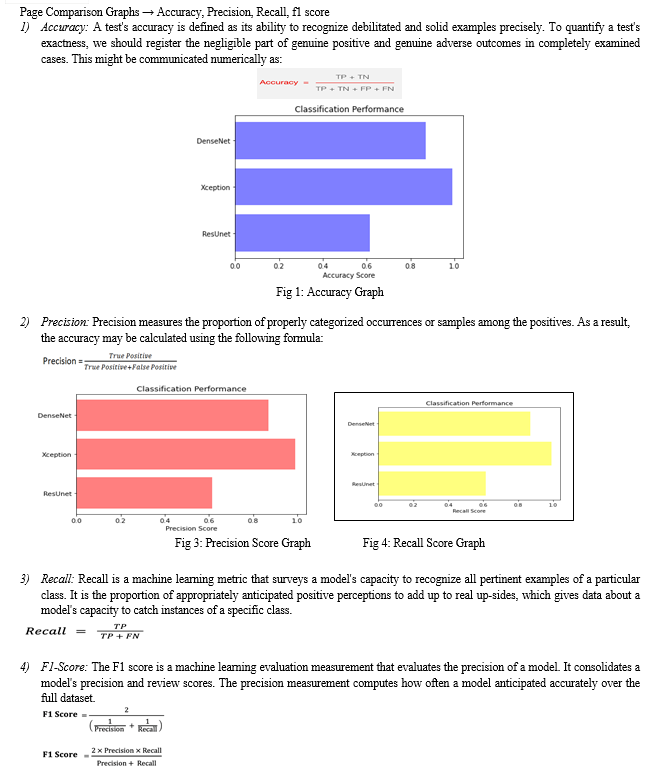
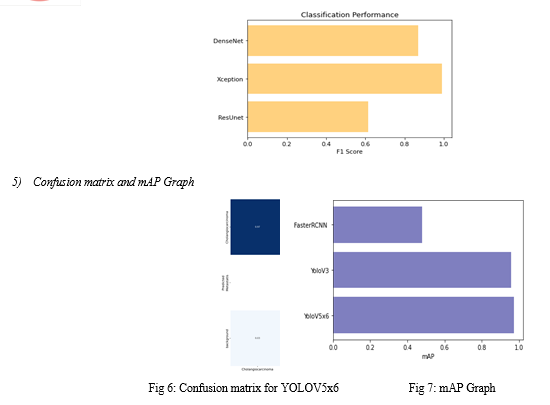
For testing, we evaluate the pre-trained ensemble model on liver tumor images not included in the training data. We compute performance metrics such as accuracy, precision, recall, and F1-score to evaluate the system's tumor diagnosis and localization capabilities.
To ensure model robustness and generalization, we may utilize cross-validation techniques across diverse datasets. Through rigorous training and testing, our system aims to showcase superior performance in liver tumor diagnosis, providing a dependable solution for early detection and management, potentially transforming global clinical practices.
IV. EXPERIMENTAL RESULTS
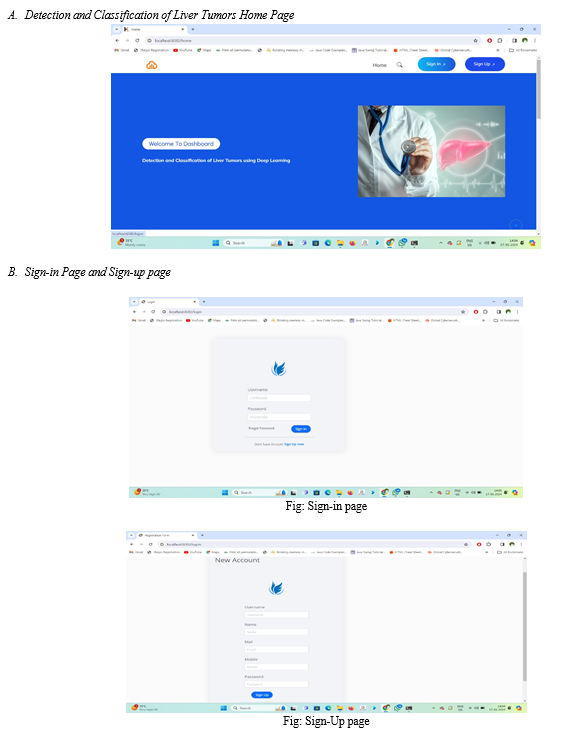
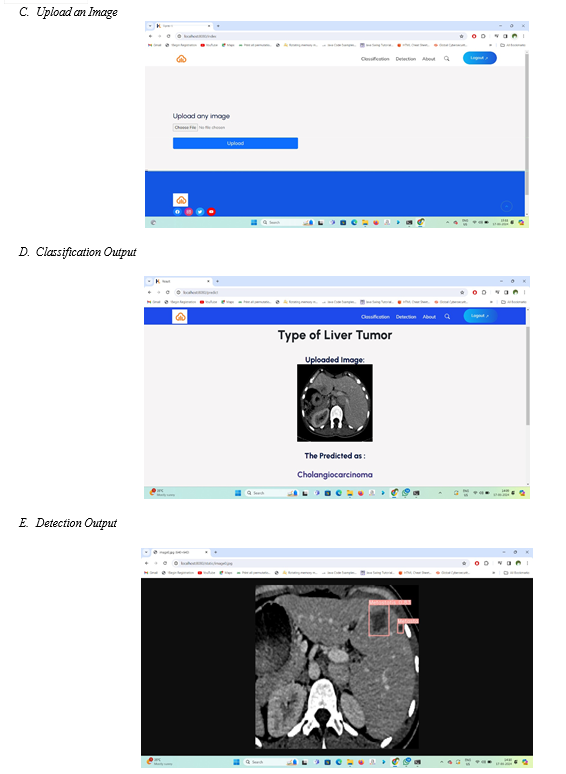
The user interface is provided for user where user has to enter the details and upload an image.
Conclusion
In conclusion, our study presents Liver ResUnet, a comprehensive framework tailored for the detection and classification of liver tumors utilizing deep learning methodologies. By incorporating various techniques including image preprocessing, data augmentation, and ensemble learning, we have developed a robust system, ResUNet, capable of accurately classifying liver tumors. Furthermore, our integration of detection models such as FasterRCNN, YoloV3, and YoloV5 enhances the framework\'s capability to accurately identify tumor regions within liver images. The implementation of Flask with SQLite for user interaction ensures ease of access and usability, allowing users to upload images for real-time analysis. Through rigorous training and optimization, our framework achieves better accuracy and efficiency in tumor detection and classification tasks. Additionally, by exploring alternative architectures such as DenseNet and Xception, we demonstrate the adaptability and scalability of our approach, further enhancing performance. Overall, Liver ResUnet presents a versatile and user-friendly solution for liver tumor analysis, with potential applications in medical diagnostics and research. Our framework not only advances the field of medical imaging but also underscores the efficacy of deep learning techniques in addressing complex healthcare challenges. Liver ResUnet\'s future development could focus on integrating multi-modal data, supporting real-time processing, incorporating interpretability techniques, adding automated reporting functionalities, seamlessly integrating with electronic health record systems, and ensuring continuous updates and fine-tuning with emerging methodologies and datasets.
References
[1] M. P. Arakeri. (2011). “Recent advances and future potential of computer aided diagnosis of liver cancer on computed tomography images,” in Proc. of Int. Conf. on Information Processing, Bangalore, India, pp. 246–25 [2] K. C. Kaluva, M. Khened, A. Kori and G. Krishnamurthi. (2018). “2D-densely connected convolution neural networks for automatic liver and tumor segmentation,” Computer Vision and Pattern Recognition, Cornell University. arXiv preprint arXiv:1800218 [3] F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre et al. (2018). , “Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries,” CA: A Cancer Journal for Clinicians, vol. 68, no. 6, pp. 394–424. [4] K. Yasaka, H. Akai, O. Abe and S. Kiryu. (2018). “Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: A preliminary study,” Radiology, vol. 286, no. 3, pp. 887–896. [5] A.Gotra, I. Sivakumaran, G. Chartrand, K. N. Vu, F. Vandenbroucke-Menu et al. (2017). , “Liver segmentation: Indications, techniques and future directions,” Insights into Imaging, vol. 8, no. 4, pp. 377–392. [6] Y. Todoroki, X. H. Han, Y. Iwamoto, L. Lin, H. Hu et al. (2017). , “Detection of liver tumor candidates from CT images using deep convoluteonal neural networks,” in Proc. of Int. Conf. on Innovation in Medicine and Healthcare, Puerto de la Cruz, Spain, pp. 140–145. [7] M. H.Forouzanfar, A. Afshin, L. T. Alexander, H. R. Anderson, Z. A. Bhutta et al. (2016). , “Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2015: A systematic analysis for the global burden of disease study 2015,” Lancet, vol. 388, no. 10053, pp. 1659–1724. [8] M. Bellver, K. K. Maninis, J. Pont-Tuset, X. Giró-i-Nieto, J. Torres et al. (2017). , “Detection-aided liver lesion segmentation using deep learning,” Computer Vision and Pattern Recognition, Cornell University. arXiv preprint arXiv: 1711.11069. [9] F. X. Bosch, J. Ribes, M. Díaz and R. Cléries. (2004). “Primary liver cancer worldwide incidence and trends,” Gastroenterology, vol. 127, no. 5, pp. S5–S16. [10] K. K. Brock. (2011). “Imaging and image-guided radiation therapy in liver cancer,” in Seminars in Radiation Oncology. vol. 21, WB Saunders, pp. 247–255. [11] S. R. Abdel-Misih and M. Bloomston. (2010). “Liver anatomy,” Surgical Clinics, vol. 90, no. 4, pp. 643–653. [12] M. Ahmad, I. Albatish, M. J. Mosa and S. S. Abu-Naser. (2017). “Knowledge based system for long-term abdominal pain (stomach pain) diagnosis and treatment,” International Journal of Engineering and Information Systems, vol. 1, no. 4, pp. 71–88. [13] M. A. Frid, I. Diamant, E. Klang, M. Amitai, J. Goldberger et al. (2018). , “GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification,” Neurocomputing, vol. 321, pp. 321–331. [14] T. Christian and E. Olsvik. (2018). “Biometric Fish Classification of Nordic Species Using Convolutional Neural Network with Squeeze-and-Excitation,” Master’s thesis, University of Agder, Universitetet i Agder. [15] X. Han. (2017). “Automatic liver lesion segmentation using a deep convolutional neural network method,” Computer Vision and Pattern Recognition, Cornell University. arXiv preprint arXiv:1704.072392017. [16] Y. LeCun, Y. Bengio and G. Hinton. (2015). “Deep learning,” Nature, vol. 521, no. 7553, pp. 436–444. [17] W. Li, F. Jia and Q. Hu. (2015). “Automatic segmentation of liver tumor in CT images with deep convolutional neural networks,” Journal of Computer and Communications, vol. 3, no. 11, pp. 146–150. [18] F. Pan, Q. Huang and X. Li. (2019). “Classification of liver tumors with CEUS based on 3D-CNN,” in Proc. 2019 IEEE 4th Int. Conf. on Advanced Robotics and Mechatronics, Toyonaka, Japan: IEEE, pp. 845–849. [19] B. Míriam, K. K. Maninis, J. P. Tuset, X. Giró-i-Nieto, J. Torres et al. (2017). , “Detection-aided liver lesion segmentation using deep learning,” Computer Vision and Pattern Recognition, Cornell University. arXiv preprint arXiv:1711.11069. [20] G. Chlebus, A. Schenk, J. H. Moltz, B. van Ginneken, H. K. Hahn et al. (2018). , “Automatic liver tumor segmentation in CT with fully convolutional neural networks and object-based postprocessing,” Scientific Reports, vol. 8, no. 1, pp. 1–7. [21] J. Son, S. J. Sang, J. Park and K. H. Jung. (2017). “Retinal vessel segmentation in fundoscopic images with generative adversarial networks,” Computer Vision and Pattern Recognition, Cornell University. arXiv preprint arXiv:1706.09318. [22] S. Almotairi, G. Kareem, M. Aouf, B. Almutairi and M. A. M. Salem. (2020). “Liver tumor segmentation in CT scans using modified signet,” Sensors, vol. 20, no. 5, pp. 1516.
Copyright
Copyright © 2024 Ajjuguttu Triveni Reddy, Dova Bhavana, Athota Venkataramaiah, Ala Gopal, Mrs. B. Lalitha Rajeswari. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET59142
Publish Date : 2024-03-19
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

