Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Dermatological Categorization Using Deep Learning
Authors: Sangmeshwar Kawdi, Vinay , Naveed
DOI Link: https://doi.org/10.22214/ijraset.2024.59523
Certificate: View Certificate
Abstract
Skin disease is a rampant medical condition in today\'s society that demands immediate attention. The manifestation of small circular or irregularly shaped spots on the skin is a clear indication of skin disease. If left untreated, skin disease could progress into skin cancer, a condition that poses a great danger to the patient\'s life. This project is set to detect possible signs of skin abrasions or infections. By utilizing neural networks or machine learning algorithms to scour an open-source dataset of skin diseases, the project provides the most accurate match to the patient\'s likely condition. We will use CNN transfer learning techniques, particularly VGG19 or different Resnet approaches, to ensure maximum accuracy. Our goal is to cover at least 12+ out of the 23 available classes and achieve an accuracy rate of 90% or higher on the training set and 85% or higher on the validation set. The project will also include the best available treatments to treat any detected disease with a tailored dataset. We are committed to providing top-notch medical care and ensuring the best possible outcome for our patients.
Introduction
I. INTRODUCTION
Our research proposes a groundbreaking approach to analyzing skin conditions using only a smartphone camera module, eliminating the need for a separate skin diagnosis device. Our method accurately detects common skin conditions such as acne, pigmentation, blemish, and flush in facial images captured by a smartphone camera. Our innovative approach employs Haar features to identify face features and regions, as well as YCbCr and HSV color models to identify skin regions. By setting the hue range of a component image, we are able to detect acne and flush, while pigmentation is calculated by finding the factor between the minimum and maximum values of the corresponding skin pixel in the component image R. Additionally, blemish is detected by applying adaptive thresholds in grayscale images. Our results demonstrate a high level of accuracy and reliability, and we are confident that our approach has immense potential in the field of skin analysis and diagnosis.
The field of skin lesion classification is a fascinating area of study within dermatoscopic lesion image processing. Our team has developed an innovative approach to skin lesion classification using a deep Convolutional Neural Network (CNN) called MobileNet. To make the model even more effective, we modified it specifically for skin lesion classification, resulting in a highly efficient and accurate tool for processing dermatoscopic lesion images. In order to evaluate our model, we utilized the Human Against Machine (HAM) dataset, which includes 10,000 training images from various sources. The HAM dataset is incredibly diverse, containing skin lesion images with varying characteristics. Our modified model outperformed the traditional MobileNet in terms of accuracy, specificity, sensitivity, and F1-score. This approach has enormous potential for accurate and efficient classification of skin lesions, making it a valuable tool in the field of dermatology.
In recent years, deep learning has emerged as a highly sought-after method and a powerful tool in complex fields that require a priori knowledge. In the field of biomedicine, where medical resources are scarce, deep learning has proven to be a promising solution for diagnosing diseases. Dermatology, in particular, has greatly benefited from the application of deep learning in the classification of skin diseases. This paper provides a comprehensive overview of the use of deep learning in classifying skin diseases. It summarizes the key characteristics of skin lesions and the current state of image technology. The paper meticulously analyzes various studies on skin disease classification using deep learning and critically examines the datasets, data processing techniques, classification models, and evaluation criteria used in these studies.
II. LITERATURE REVIEW
Several machine learning techniques have been utilized by authors to classify skin diseases. While support vector machine, artificial neural network, and decision trees are widely used, deep learning-based approaches have gained popularity due to their high accuracy and compatibility with image inputs. This literature survey compares traditional classification methods with deep learning- based approaches, and also explores transfer learning and ensemble learning.
Previously common techniques like support vector machine and artificial neural networks have been surpassed by convolutional neural network- based models, AlexNet models, ResNet 50/ResNet 101, and dense CNN models. Transfer learning is also gaining attention, where deep learning models are used to extract features for other models. Bashar A. conducted a survey on neural network deep learning techniques and their applications, while Raj S.J. explored the limitations and benefits of machine learning methods and concluded that deep learning and optimization techniques significantly improve classification accuracy.
III. SCOPE OF WORK
Our goal is to advance research in deep learning for dermatological image analysis and to develop highly accurate and efficient diagnosis algorithms
IV. METHODOLOGY
In the field of scientific research, online datasets have emerged as a valuable resource for experiments, with several datasets such as ISIC, PH2, and EDRA being commonly used by researchers. For the current study, we have selected the ICIS public dataset, which is a comprehensive collection of over 10,000 images of melanoma and benign skin diseases. However, the ICIS dataset suffers from a data imbalance issue, which can lead to inaccurate results. To address this issue, we employed data balancing techniques before utilizing the dataset for our experiment. Notably, the benign images in the dataset represent early-stage skin cancer, while the melanoma images represent later stages of the disease. To create an experimental dataset, we randomly selected 20% of the images from both classes, resulting in a balanced dataset of 1000 images of each class. In this study, we employed a powerful deep learning model, specifically the CNN model of 2016, for feature extraction from the input images. The CNN model was capable of extracting 1000 features from each image, and manual labeling was performed for all the images. The extracted features and their corresponding labels were stored in a text file, facilitating the application of any supervised machine learning algorithm for classification. We employed various machine learning algorithms, including support vector machine, decision tree, linear regression, and K-Nearest Neighbor algorithm, for classification. The performance of each classifier was evaluated using the confusion matrix and ROC curve, providing a comprehensive analysis of their effectiveness. The input images for this study were obtained from several reputable databases, ensuring a diverse and representative sample. The images contained two types of skin diseases: benign and malignant. The benign images represented early-stage skin cancer, while the malignant images represented later stages of the disease. The dataset included thousands of images of both classes, providing a vast and comprehensive collection for experimentation.
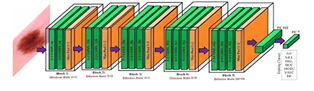
For feature extraction, we utilized the VGG 16 architecture of the CNN model, which is a well-established technique for this purpose. The VGG 16 model comprises 3 convolution layers with one pooling layer, and this combination is repeated 4 times to construct a 16-layer architecture. Each convolution layer comprises filters of 3x3 size at each layer, and the pooling layer is employed to reduce the feature size obtained from the convolution layer. The filters were used to extract low-level, mid-level, and high-level features from the image, providing a comprehensive and detailed analysis of the input data. In this study, we also employed transfer learning, which is a powerful and versatile technique that allows different domains, tasks, and distributions to be employed during training and testing. We used a transfer learning model with two distinct levels of setup, allowing for a comprehensive and accurate analysis of the input data. The first setup was used to extract low-level, mid-level, and high-level features from the input dataset using the VGG-16 model. In the second setup, we inserted labels into the features extracted from the various input images of stage 1, providing a comprehensive and detailed analysis of the input data. Finally, all the features were sent to the supervised machine learning model for classification, providing a powerful and accurate analysis of the input data. We employed classifiers, including support vector machine, decision tree, KNN, and Linear Discriminant analyzer, as they are suitable for binary and linear classification, providing a powerful and accurate analysis of the input data. Overall, this study provides a comprehensive and detailed analysis of the input data, employing powerful and versatile techniques to provide accurate and informative results. VGG19 and VGG16 are both deep convolutional neural network architectures proposed by the Visual Geometry Group (VGG) at the University of Oxford. The main difference between the two lies in their depth: VGG19 has 19 layers (hence the name) while VGG16 has 16 layers, In summary, VGG19 is more advanced than VGG16 primarily due to its deeper architecture, which allows it to capture more complex features from the data. However, this increased depth also comes with higher computational costs and the risk of overfitting, highlighting the trade-offs involved in designing and using deep neural networks. Hence we use VGG19 architecture.
Our models come with pre-trained weights that are automatically downloaded when you instantiate a model. And with our models, you can perform tasks like prediction, feature extraction, and fine-tuning. Plus, our models are built to match the image data format set in your Keras configuration file, ensuring seamless integration with your existing workflow.
Conclusion
To achieve optimal classification accuracy up to the signature mark, the proposed work relies on transfer learning. It\'s worth noting that the pure deep learning model falls short in comparison to the transfer learning model. While the transfer learning model may require longer execution time and human intervention for data labeling, it still outperforms deep learning models. In our experiments, we consistently achieved over 99% accuracy with models such as decision trees and K nearest neighbors. However, complex models like ensemble learning with boosted trees showed inadequate performance, with less than 50% accuracy. Therefore, it\'s important to recognize that the above dataset is highly suitable for linear binary classifiers and not appropriate for coarse multiclass models.
References
[1] Haenssle, Holger A., et al. \"Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists.\" Annals of Oncology 29.8 (2018): 1836-1842. [2] Walker, B. N., et al. \"Dermoscopy diagnosis of cancerous lesions utilizing dual deep learning algorithms via visual and audio (sonification) outputs: Laboratory and prospective observational studies.\" EBioMedicine 40 (2019): 176-183. [3] Brinker, Titus Josef, et al. \"Skin cancer classification using convolutional neural networks: systematic review.\" Journal of medical Internet research 20.10 (2018): e11936. [4] Maron, Roman C., et al. \"Systematic outperformance of 112 dermatologists in multiclass skin cancer image classification by convolutional neural networks.\" European Journal of Cancer 119 (2019): 57-65. [5] Tschandl, Philipp, et al. \"Expert -level diagnosis of nonpigmented skin cancer by combined convolutional neural networks.\" JAMA dermatology 155.1 (2019): 58-65. [6] Xue, Cheng, et al. \"Robust Learning at Noisy Labeled Medical Images: Applied to Skin Lesion Classification.\" arXiv preprint arXiv:1901.07759 (2019). [7] He, Xinzi, et al. \"Dense deconvolution net: Multi path fusion and dense deconvolution for high resolution skin lesion segmentation.\" Technology and Health Care 26.S1 (2018): 307-316. [8] Burdick, Jack, et al. \"Rethinking skin lesion segmentation in a convolutional classifier.\" Journal of digital imaging 31.4 (2018): 435-440. [9] Brinker, Titus J., et al. \"Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task.\" European Journal of Cancer 113 (2019): 47-54. [10] Zhang, Xinyuan, et al. \"Towards improving diagnosis of skin diseases by combining deep neural network and [11] human knowledge.\" BMC medical informatics and decision making 18.2 (2018): 59. [12] Hekler, Achim, et al. \"Pathologist-level classification of histopathological melanoma images with deep neural networks.\" European Journal of Cancer 115 (2019): 79-83. [13] Tschandl, Philipp, Christoph Sinz, and Harald Kittler. \"Domain-specific classification-pretrained fully convolutional network encoders for skin lesion segmentation.\" Computers in biology and medicine 104 (2019): 111-116. [14] Mahbod, Amirreza, et al. \"Skin lesion classification using hybrid deep neural networks.\" ICASSP 2019-2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). IEEE, 2019. [15] Marchetti, Michael A., et al. \"Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images.\" Journal of the American Academy of Dermatology 78.2 (2018): 270-277. [16] Chaturvedi, Saket S., Kajol Gupta, and Prakash Prasad. \"Skin Lesion Analyser: An Efficient Seven-Way Multi-Class Skin Cancer Classification Using MobileNet.\" arXiv preprint arXiv:1907.03220 (2019). [17] Hosny, Khalid M., Mohamed A. Kassem, and Mohamed M. Foaud. \"Classification of skin lesions using transfer learning and augmentation with Alex-net.\" PloS one 14.5 (2019): e0217293 [18] Bisla, Devansh, et al. \"Towards Automated Melanoma Detection With Deep Learning: Data Purification and Augmentation.\" Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops. 2019. [19] . Zhang, Jianpeng, et al. \"Attention residual learning for skin lesion classification.\" IEEE transactions on medical imaging (2019). [20] . Shi, Xueying, et al. \"An Active Learning Approach for Reducing Annotation Cost in Skin Lesion Analysis.\" arXiv preprint arXiv:1909.02344 (2019). [21] . Khan, Muhammad Attique, et al. \"Multi-Model Deep Neural Network based Features Extraction and Optimal Selection Approach for Skin Lesion Classification.\" 2019 International Conference on Computer and Information Sciences (ICCIS). IEEE, 2019.
Copyright
Copyright © 2024 Sangmeshwar Kawdi, Vinay , Naveed . This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET59523
Publish Date : 2024-03-28
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

