Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Designing Potential Antisense Oligonucleotides (ASO) against Lung Cancer Associated microRNA and Studying their Interaction
Authors: Indra Sarkar, Suvankar Karmakar, Santi Ranjan Dey, Samiran Mondal
DOI Link: https://doi.org/10.22214/ijraset.2024.58379
Certificate: View Certificate
Abstract
MicroRNAs (miRNAs) are an abundant class of small non-protein-coding RNAs that function as gene regulators. They regulate diverse biological processes and bioinformatics data indicates that each miRNA can control hundreds of gene targets, underscoring the potential influence of miRNAs on almost every genetic pathway. miRNA mutations or mis-expression correlate with various human cancers and miRNAs can function as tumor suppressors and oncogenes. miRNAs have been shown to repress the expression of important cancer-related genes and might prove useful in the diagnosis and treatment of cancer. In this context, we identified various miRNAs associated with the prevalent cancer phenotype NSCLC/SCLC and used the respective target miRNA sequences as templates for designing antisense oligonucleotides (ASOs). We investigated the interaction of this cognate oligonucleotides and target miRNAs to hypotheses the inhibitory potential of such oligonucleotides through molecular docking studies.
Introduction
I. INTRODUCTION
Cancer become one of the leading causes of death globally, accounting for an estimated 10 million deaths, and International Agency for Research on Cancer (IARC) provided estimates of 2.21 million new cases and 1.8 million lung-cancer related deaths in the world in 2020 [https://gco.iarc.fr]. Primary lung cancer arises from the epithelial cells, the cells lining the larger and smaller airways (bronchi and bronchioles); for this reason, lung cancers are sometimes called bronchogenic cancers or bronchogenic carcinomas. Based on histology, lung cancers are divided into non–small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) [1]. NSCLC, usually starts in glandular cells on the outer part of the lung. This type of cancer is called adenocarcinoma. NSCLC can also start in flat, thin cells called squamous cells. These cells line the bronchi, which are the large tubes, or airways, that branch off from the trachea, or windpipe, into the lungs. Large cell carcinoma is another type of non–small cell lung cancer, but it is less common. There are also several rare types of non–small cell lung cancer. These include sarcoma and sarcomatoid carcinoma. NSCLC represents about 80-85% of all lung cancers and leading cause of cancer death according to the World Health Organization (WHO) [2]. SCLC, usually starts in cells that line the bronchi in the centre of the lungs. The main types of small cell lung cancer are small cell carcinoma and combined small cell carcinoma (mixed tumour with squamous or glandular cells). SCLC comprises 15-20% of all lung cancer cases [2].
MiRNAs are non-coding, single-stranded RNAs of ~20-24 nucleotides and constitute a novel class of gene regulators that are found in both plants and animals [3]. They negatively regulate their targets in one of two ways depending on the degree of complementarity between the miRNA and the target [4]. miRNAs that bind with perfect or nearly perfect complementarity to protein coding mRNA sequences induce the RNA-mediated interference (RNAi) pathway. Briefly, mRNA transcripts are cleaved by ribonucleases in the miRNA-associated, multi-protein RNA induced-silencing complex (RISC), which results in the degradation of target mRNAs [5]. This mechanism of miRNA-mediated gene silencing is commonly found in plants, but miRNA-directed mRNA cleavage has also been shown to occur in mammals [6]. However, most animal miRNAs are thought to use a second mechanism of gene regulation that does not involve the cleavage of their mRNA targets. These miRNAs exert their regulatory effects by binding too imperfect complementary sites within the 3′-untranslated regions (3′-UTRs) of their mRNA targets, and they repress target-gene expression post-transcriptionally, apparently at the level of translation, through a RISC complex that is like, or possibly identical with, the one that is used for the RNAi pathway [7].
Consistent with translational control, miRNAs that use this mechanism reduce the protein levels of their target genes, but the mRNA levels of these genes are barely affected.
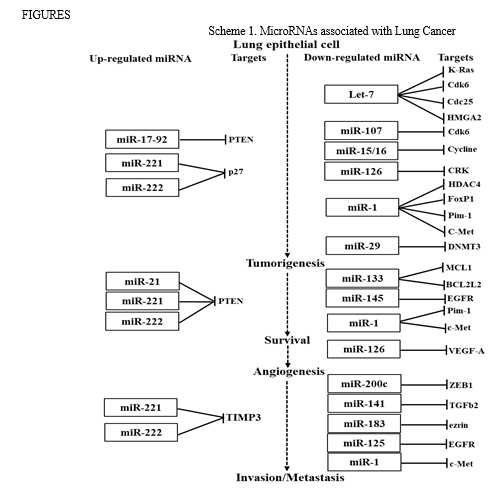
Recently, there is increasing evidence to suggest that miRNAs play important and complex roles in human cancers, including lung cancer [8-9]. The mechanisms of cell development, cell proliferation, invasion, migration, apoptosis, and metastasis in NSCLC and SCLC both have been shown in numerous studies to be significantly influenced by miRNAs [10-11] and have been identified as essential and critical regulators of biological functions as tumour suppressors or oncogenes (oncomiRs) in lung cancer. Up-regulation of oncomiRs potentially repressed the tumour suppressor genes which are responsible for tumour cell growth and also increases cell proliferation, migration and invasion. Tumor suppressor miRNAs, on the other hand, suppressed the translation of oncogenic mRNAs. Therefore, down-regulation of tumour suppressor miRNAs are responsible for tumourigenesis and development of cancers. miR-21 and miR-17-92 cluster causing tumorigenesis, are commonly upregulated in several types of cancers including NSCLC by downregulating tumour suppressor phosphate and tensin homolog (PTEN) [12-13]. The expression of the members of miR-34 family, miR-34a, miR-34b and miR-34c were downregulated in lung cancers. In SCLC tumors, miR-34 family members are thought to act as a tumour suppressor [14]. Upregulation of miR-221 and miR-222 was manifested in different types of tumour involve in cell growth and invasion by targeting PTEN and TIMP3 [15]. On the other side, a tumour suppressor miRNA, let-7 was reported to reduce in NSCLC induces the downregulation of KRAS [16]. Numerous studies have reported the oncogenic and tumour suppressor activity of miRNAs associated with lung cancers, specifically in case of NSCLC are summarizing in schematic diagram presented in Scheme 1.
For NSCLC patients, chemotherapy is the main therapies having lack of effectiveness and selectivity for tumour cells. Therefore, new effective and targeted therapeutics involving antisense oligonucleotides (ASOs) represent a promising alternative by using this class of molecules as inhibitors of specific miRNA, showing an improved specificity compared to traditional anti-cancer drugs. In this context, the primary objective of the current study is to identify various miRNAs associated with the prevalent cancer phenotype NSCLC/SCLC and segregated them into up-regulated and down-regulated categories. We used the respective target miRNA sequences as templates for designing antisense oligonucleotides (ASOs) and also generate 3D structures of the sequences. Finally, we performed molecular docking study to investigate the interaction of this cognate oligonucleotides and target miRNAs to hypotheses the inhibitory potential of such oligonucleotides.
II. MATERIALS AND METHODS
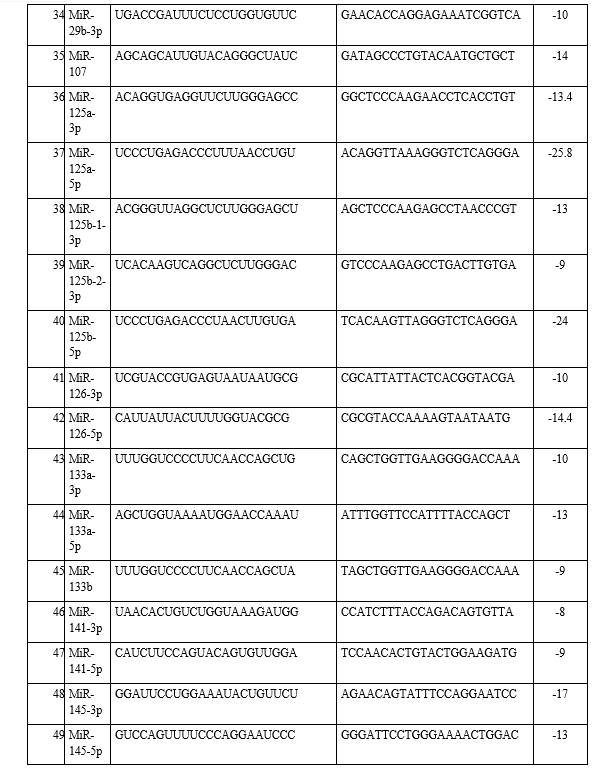
The microRNA sequences associated with lung cancer (NSCLC/SCLC) were downloaded from miRBase after identifying them from Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database [17]. s-fold is a software (https://sfold.wadsworth.org/cgi-bin/soligo.pl) is used for the target accessibility prediction and rational design of antisense oligonucleotides. For prediction of target accessibility, a complete probability profile of single-stranded regions is generated for the entire target RNA. Sites with high probabilities of being single-stranded are predicted to be accessible. At nucleotide position i, the profile shows the probability that nucleotides i, i + 1, i + 2,…,i + W − 1 are all unpaired. In other words, the profile is for consecutive fragments with a width W. Certain empirical rules of design of antisense molecules were used:(i) siRNA duplexes should be composed of 21 nt sense and 21 nt antisense strands, paired so as to each have a 2 nt 3′ dTdT overhang; (ii) the siRNA sequence should have low to moderate GC content; (iii) sequences with more than three Gs or three Cs in a row should be avoided, because polyG and polyC sequences can form quartets and may interfere in the siRNA silencing mechanism; (iv) AAAA or TTTT should also be avoided for RNA polymerase III mediated promoters because transcription tends to terminate at these sequences. It is of note that there is a lack of support in the literature for the significance of target patterns such as AA(N19) or NA(N19). The file oligo_f.out gives filtered output for antisense oligos of user-specified length. Soligo currently uses the following filters: antisense oligo binding energy ≤−8 kcal/mol;40% ≤ GC % ≤ 60%; No GGGG in the target sequence. The antisense oligo binding energy is a weighted sum of the DNA/RNA stacking energies for the hybrid formed by the antisense oligo and the targeted sequence. For a base-pair stack, the weight for the sum is calculated by the probability of the unpaired dinucleotide in the target sequence that is involved in the stack. This weighting scheme accounts for the structural variation at the target site. The 3D structures of the target (miRNA) and its antisense molecule were generated using the RNA composer server (https://rnacomposer.cs.put.poznan.pl/). The building starts from the 3D structure element which bears both 5′- and 3′-terminal residues of the target RNA structure. The Kabsch algorithm for least-squares superposition is implemented to overlay 3D structure elements, based on common canonical base pair residues [18]. Subsequently, all atom coordinates within the newly added fragment are removed to avoid the coordinate duplication.
After the addition of the last structure element, due to the graph representation, all residues are renumbered according to their sequence. Two energy minimization steps are conducted to refine the initial RNA structure. In the first step, the atom coordinates are strictly converted to the CYANA program format and energy is minimized in the torsion angle space using the standard protocol (conjugate gradient, 2000 iteration steps), as well as the distance restrains for hydrogen bonds. In the second step, the resultant structure is energy minimized in the Cartesian atom coordinate space using CHARMM force field implemented in the XPLOR-NIH program (conjugate gradient, 1000 iterations) and restraints for hydrogen bonds and base pairs planarity. RNA 3D structure quality assessment: The stereochemical quality of the predicted 3D structures was assessed using PDB validation tool http://deposit.pdb.org/adit/, X-PLOR program [18] and MolProbity tool [19]. The accuracy of the predicted full atomic structures relative to the respective RNA crystal structures is described using two measures. The global and local RNA structure all atoms R.M.S.D. values were computed using XPLOR-NIH program. The interaction network fidelity (INFall) measure [20] was used to check all the canonical and non-canonical base pairing and stacking. Base pair interactions and the base stacking network for the tertiary structures were obtained using RNAView [21] and MC-Annotate [22] programs, respectively. In a similar fashion, we have calculated the parameter INFcbp to inspect the conservation of canonical base pairs [17]. The complexes were docked using HEX standalone tool & studied their interactions using DSSR server.
III. RESULTS AND DISCUSSION
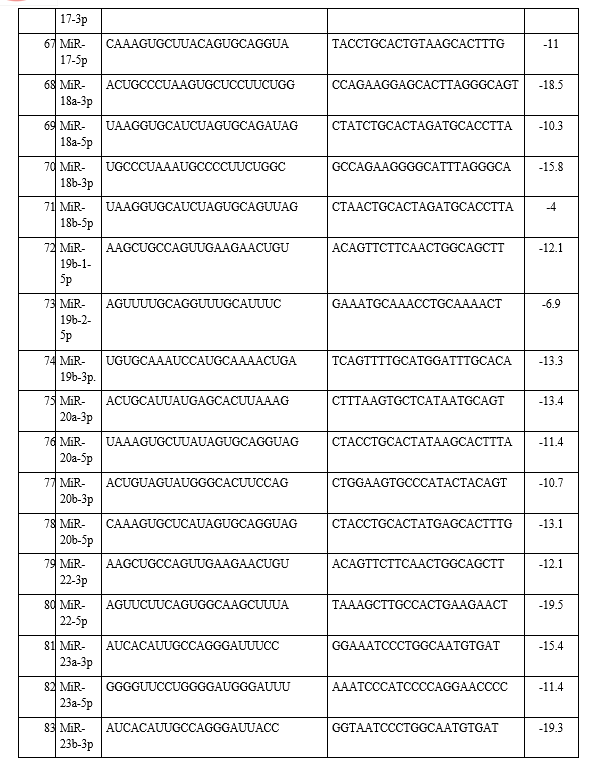
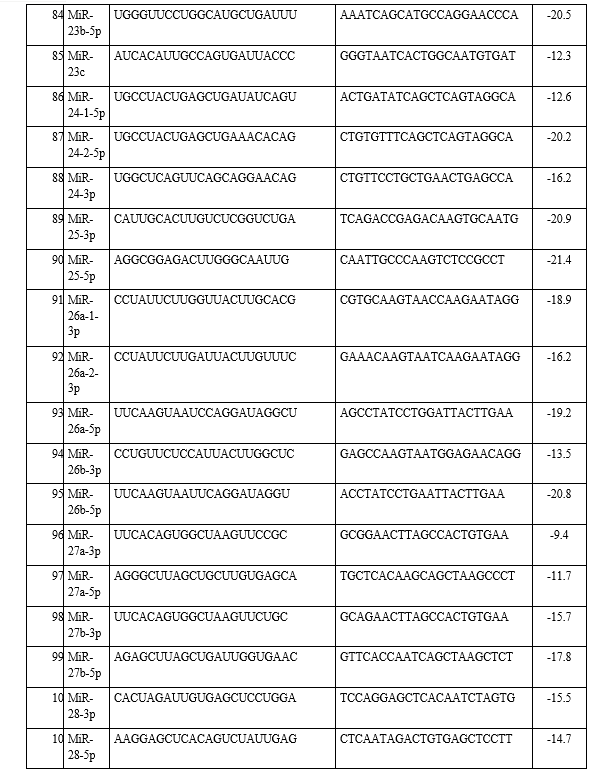
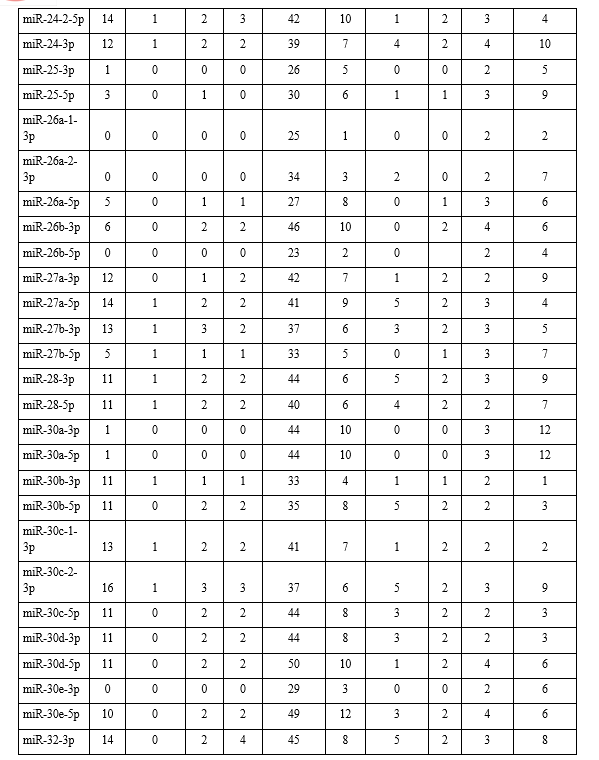
In the present study, the emerging field of ‘oncomiRs’-miRNAs that are associated with lung cancer and how they interact with each other was investigated. A total of 23 types of different miRNAs were identified from the KEGG pathway and antisense oligonucleotides of each miRNA were generated by using s-fold, corresponding to NSCLC/SCLC, are shown in Table 1. Present study involved the investigation, identification and pathway analysis of up-regulated and down-regulated miRNAs associated with lung cancers. Then antisense oligonucleotides of each miRNA were analyzed and generated the 3D structure of target and its antisense molecule.
- Interaction Of Downregulated Mirnas With Their Corresponding Antisense Oligonucleotides
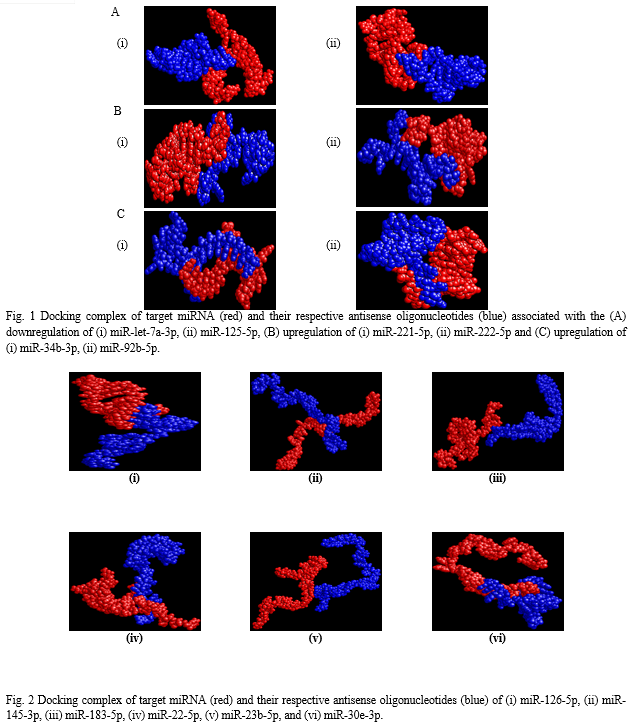
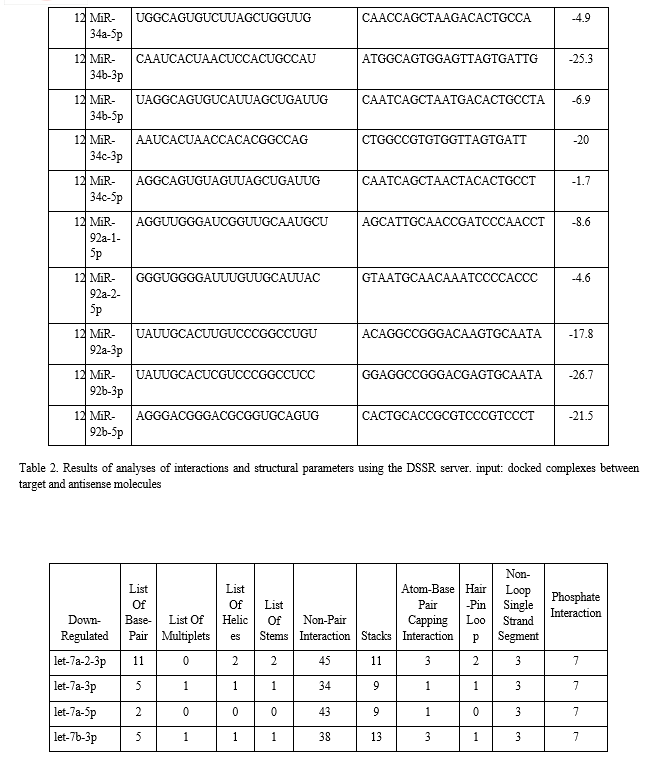
The binding energies from the docking interaction studies of each downregulated target miRNAs (as receptor) and their respective antisense oligonucleotides (as ligand) are shown in Table 1. It is evident that antisense oligonucleotides of miR-let-7, miR-125, miR-34b and miR-92b family interacted more strongly (ΔG values more negative) compared to the other members of this group. Among the miR-let-7 family members, miR-let-7b-5p (ΔG= -26 kcal mol−1) and in case of miR-125 members, miR-125a-5p (ΔG= -25.8 kcal mol−1), showed the highest interaction with their corresponding antisense oligonucleotides. Representative docking complexes of miR-let-7b-5p and miR-125a-5p with their respective antisense oligonucleotides are showing in Fig. 1(A). Both the miR-let-7 and miR-125a-5p has been reported to act as tumour suppressor in NSCLC and functionally inhibits the two main characterized oncogenes, Ras and EGFR [23-25], shown in Scheme 1. Results of analyses of interactions and structural parameters for docked complexes between target and antisense molecules using the DSSR server of the let-7 family members is shown in Table 2.
2. Interaction Of Upregulated Mirnas With Their Corresponding Antisense Oligonucleotides
The docking interaction studies of upregulated target miRNAs (as receptor) and their respective antisense oligonucleotides (as ligand) for both NSCLC and SCLC are shown in Table 1. The binding energy values for miR-221-5p and miR-222-5p were obtained as -15 and 24 kcal mol−1 respectively. Representative docking complexes of miR-221-5p and miR-222-5p with their respective antisense oligonucleotides are showing in Fig. 1(B).
Both of them are upregulated in case of NSCLC/SCLC and associated with the downregulation of p27, PTEN and TIMP3 [26-27], as shown in Scheme 1. However, in case of SCLC, miR-92b-3p was showing highest binding efficiency (ΔG= -26.7 kcal mol−1) with its antisense oligonucleotide [Table 1]. Previous report suggests that the expression of miR-92b-3p in plasma exosomes were increased to promote cell migration and chemoresistance through the PTEN/AKT pathway in SCLC patients [28]. Representative docking complexes of miR-34b-3p and miR-92b-5p with their respective antisense oligonucleotides are showing in Fig. 1(C).
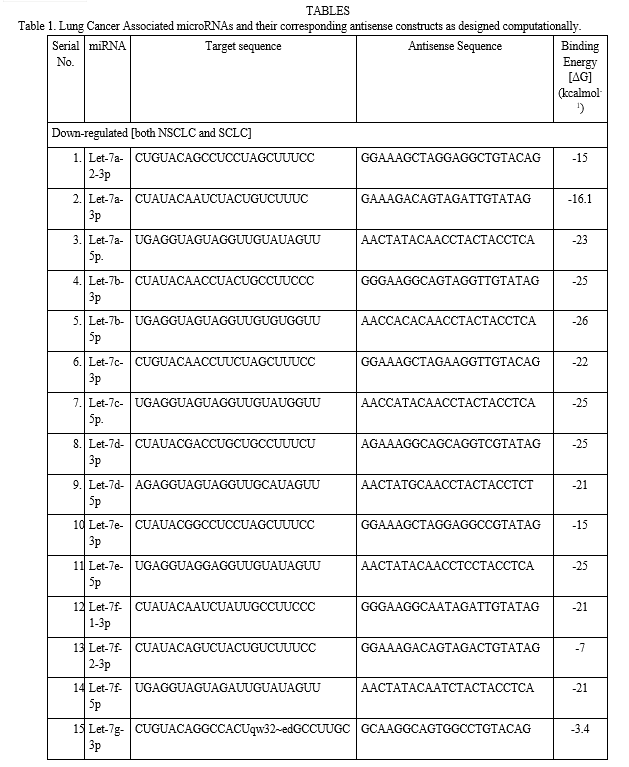
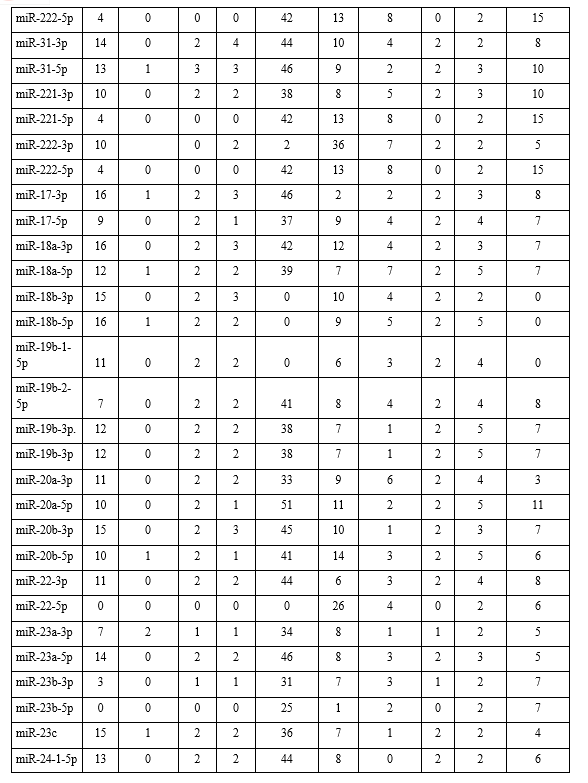
From CYANA programme it has been determined that the average binding energies for our studied 35 types of miRNA-antisense oligonucleotide associated interactions were calculated to be -63.4 kcal mol−1 (-2218.84/35 kcal mol−1) for lung cancer (Table 3). These values indicate that the interactions were stable. From the docking interaction study of each target miRNA (as receptor) and their respective antisense (as ligand), interestingly we found that 6 of the miRNA-antisense (miR-126-5p, miR-145-3p, miR-183-5p, miR-22-5p, miR-23b-5p, and miR-30e-3p) interactions were devoid of hydrogen bond, showing in Table 2. However, as is evident from the results, the non-bonded interactions stabilized the modalities of the association of the two molecules, showing in Fig. 2.
Finally, tables (Table 2) were created where number of base pair, multiplets, helices, stems, non-pair interactions, stacks, atom-base capping interaction, hair-pin loop, non-loop single stranded segment, phosphate interaction of each complex were listed to show the types and levels of interactions.
IV. ACKNOWLEDGEMENTS
This work was supported by DSTBT, GoWB, India [Grant No. 1352(Sanc.)/STBT-11012(25)/46/2021-ST-SEC to S.M.] and Research Fund grant [R21-3846200522], the Royal Society of Chemistry, UK. S.K. thank DSTBT, GoWB for his fellowships. S.M. acknowledges Rammohan College, Kolkata for infrastructural facilities.
Conflict of Interest
The authors declare no competing financial interests.
Declarations
Ethical Approval
Not applicable.
Availability of data and materials
Authors will provide all data and materials on contact directly to authors.









Conclusion
As it is evident from the results, the designing of the antisense oligonucleotides against the target miRNAs were successful since the free energy/relative entropy calculated for the docking runs were all negative as well as displayed a uniform pattern dependent on miRNA type. Out of hundreds of miRNAs that have recently been identified, only a small number from worm, fly and human genomes have been characterized. They have been shown to control cell growth, differentiation and apoptosis; consequently, impaired miRNA expression has been implicated in tumorigenesis. Continued research into miRNA function might lead to an advanced understanding of the mechanisms that lead to tumorigenesis. It is believed that these designed antisense constructs can be tested in the wet lab for their efficacy of binding the cognate miRNAs and further be developed into effective therapeutic interventions for the treatment or prevention of cancer progression.
References
[1] Iqbal MA, Arora S, Prakasam G, Calin GA, Syed MA. MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med, 2019, 70, 3-20. [2] Sher T, Dy GK, Adjei AA. Small cell lung cancer. Mayo Clin Proc, 2008, 83, 355-367. [3] Ying SY, Chang DC, Lin SL. The microRNA (miRNA): overview of the RNA genes that modulate gene function. Mol Biotechnol. 2008 Mar;38(3):257-68. [4] Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010 Nov;11(7):537-61. [5] Van den Berg A, Mols J, Han J. RISC-target interaction: cleavage and translational suppression. Biochim Biophys Acta. 2008 Nov;1779(11):668-77. [6] Millar AA, Waterhouse PM. Plant and animal microRNAs: similarities and differences. Funct Integr Genomics. 2005 Jul;5(3):129-35. doi: 10.1007/s10142-005-0145-2. [7] Cannell IG, Kong YW, Bushell M. How do microRNAs regulate gene expression? Biochem Soc Trans. 2008 Dec;36(Pt 6):1224-31. doi: 10.1042/BST0361224. [8] Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM: A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA, 2006, 103, 2257–2261. [9] Hui A, How C, Ito E, Liu FF: Micro-RNAs as diagnostic or prognostic markers in human epithelial malignancies. BMC Cancer 2011, 11, 500. [10] Zhong S, Golipon H, Zardo P et al, miRNAs in lung cancer. A systematic review identifes predictive and prognostic miRNA candidates for precision medicine in lung cancer. Transl Res, 2021, 230, 164-196. [11] Azizi MIHN, Othman I, Naidu R, The Role of MicroRNAs in Lung Cancer Metabolism, Cancers, 2021, 13, 1716. [12] Y. Huang, Y. B. Yang, X. H. Zhang, X. L. Yu, Z. B. Wang, and X. C. Cheng, “MicroRNA-21 gene and cancer,” Medical Oncology, vol. 30, no. 1, p. 376, 2013. [13] C. P. Concepcion, C. Bonetti, and A. Ventura, “The microRNA-17-92 family of microRNA clusters in development and disease,” 5e Cancer Journal, vol. 18, no. 3, pp. 262-267, 2012. [14] Mizuno K, Mataki H, Atsushi T et. al. (2017) The microRNA expression signature of small cell lung cancer: tumor suppressors of miR-27a-5p and miR-34b-3p and their targeted oncogenes. J Hum Genet 62(7):671–678. [15] Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, Gasparini P, Gonelli A, Costinean S, Acunzo M, Condorelli G, Croce CM. Retraction Notice to: miR-221&222 Regulate TRAIL Resistance and Enhance Tumorigenicity through PTEN and TIMP3 Downregulation. Cancer Cell. 2022 Nov 14;40(11):1440. [16] Saridaki Z, Weidhaas JB, Lenz HJ, Laurent-Puig P, Jacobs B, De Schutter J, De Roock W, Salzman DW, Zhang W, Yang D, Pilati C, Bouché O, Piessevaux H, Tejpar S. A let-7 microRNA-binding site polymorphism in KRAS predicts improved outcome in patients with metastatic colorectal cancer treated with salvage cetuximab/panitumumab monotherapy. Clin Cancer Res. 2014 Sep 1;20(17):4499-4510. [17] Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016 Jan 4;44(D1): D457-62. [18] Popenda M, Szachniuk M, Antczak M, Purzycka KJ, Lukasiak P, Bartol N, Blazewicz J, Adamiak RW. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012 Aug;40(14): e112. [19] I. W. Davis, A. Leaver-Fay, V. B. Chen, J. N. Block, G. J. Kapral, X. Wang, L. W. Murray, W. B. Arendall, III, J. Snoeyink, J. S. Richardson, and D. C. Richardson. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids, Nucl. Acids Res. 35: W375-W383 (2007). [20] Parisien,M., Cruz,J.A., Westhof,E. and Major,F. (2009) New metrics for comparing and assessing discrepancies between RNA 3D structures and models. RNA, 15, 1875–1885. [21] Yang H, Jossinet F, Leontis N, Chen L, Westbrook J, Berman H, Westhof E: Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res 2003, 31: 3450-3460. [22] Gendron,P., Lemieux,S. and Major,F. (2001) Quantitative analysis of nucleic acid three-dimensional structures. J. Mol. Biol., 308, 919-936. [23] Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA 2008, 105, 3903-3908. [24] Amri J, Molaee N, Karami H. Up-Regulation of MiRNA-125a-5p Inhibits Cell Proliferation and Increases EGFR-TKI Induced Apoptosis in Lung Cancer Cells. Asian Pac J Cancer Prev. 2019 Nov 1;20(11):3361-3367. [25] Jiang, L., Huang, Q., Zhang, S. et al. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer 10, 318-330 (2010). [26] Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, Gasparini P, Gonelli A, Costinean S, Acunzo M, Condorelli G, Croce CM. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009 Dec 8;16(6):498-509. [27] Li M, Shan W, Hua Y, Chao F, Cui Y, Lv L, Dou X, Bian X, Zou J, Li H, Lin W. Exosomal miR-92b-3p Promotes Chemoresistance of Small Cell Lung Cancer Through the PTEN/AKT Pathway. Front Cell Dev Biol. 2021 May 31;9:661602.
Copyright
Copyright © 2024 Indra Sarkar, Suvankar Karmakar, Santi Ranjan Dey, Samiran Mondal. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET58379
Publish Date : 2024-02-09
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

