Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Diagnosis of Skin Cancer using Deep Learning
Authors: Sharadindu Adhikari, Tanmay Kumar Agrawal, Soumyadip Mondal, Ayushmaan Agarwal
DOI Link: https://doi.org/10.22214/ijraset.2024.64032
Certificate: View Certificate
Abstract
Early detection of skin cancer, including types such as basal cell carcinoma and melanoma, is crucial for effective treatment and reducing mortality rates. Skin cancer diagnoses often surpass those of all other cancers combined, with rising global mortality rates underscoring the need for improved detection methods. Early identification significantly enhances treatment outcomes, as confirmed by the World Health Organization. Advances in image processing and machine vision, particularly through convolutional neural networks (CNNs), have shown that deep learning models can outperform experienced dermatologists in skin cancer detection. In this project, we developed a deep learning model using Python libraries to categorize dermoscopic images from the MNIST HAM-10000 dataset into malignant or benign categories. The model\'s performance was further evaluated against three datasets from the International Skin Imaging Collaboration (ISIC) challenge archives, with the goal of enhancing early detection and treatment of skin cancer to lower mortality rates.
Introduction
I. INTRODUCTION
Before the 1980s, melanoma detection relied on identifying macroscopic features, which were often only noticeable when the melanoma was already large, leading to delayed detection and rising mortality rates. In 1985, researchers at New York University introduced the ABCD acronym (Asymmetry, Border irregularity, Color variegation, Diameter) to help the public recognize melanoma early. After 1990, physician-led screenings and full-body imaging became standard, with computer-aided digital image analysis later emerging as a more sensitive and specific detection method. This shift motivated our work, despite not being medical students.
Skin cancer rates are rising due to pollution and climate change, with two main types: melanoma and non-melanoma. UV radiation, intensified by ozone layer depletion, is a key cause. A 10% ozone depletion is predicted to cause 3 million new non-melanoma and 4,500 melanoma cases annually [6]. Melanoma is more dangerous and prevalent in white people, with 100,350 new cases in the U.S. this year—60,190 men and 40,160 women [5]. It is the 5th most common cancer in men and 6th in women [5]. Melanoma primarily affects older adults but is increasingly seen in younger people. Early detection significantly improves survival, with a 95% five-year survival rate if caught early [5]. Non-melanoma skin cancer is more common and generally curable, with over 3 million cases annually in the U.S. [5]. Basal cell carcinoma is the most prevalent, accounting for over 80% of non-melanoma cases [5]. Although non-melanoma deaths are declining due to better detection and treatment, nearly 2,000 people die yearly from basal and squamous cell carcinoma [6].
II. LITERATURE SURVEY
A. Literature Review
Recent advancements in image processing and deep learning have led to significant improvements in the early detection and classification of skin cancer. Various methods have been developed to optimize the performance of convolutional neural networks (CNNs) for this purpose. For instance, one approach utilized an improved whale optimization algorithm to enhance CNNs, resulting in superior performance compared to other methods when tested on two different datasets [1]. Similarly, a meta-heuristic optimized CNN was applied to pre-trained network models for classifying skin cancer images, demonstrating better performance than 10 popular classifiers on the DermIS and Dermquest datasets [2].
Another notable contribution is the AnResNet model, which was fine-tuned using 19,398 images and achieved high accuracy in classifying 12 different skin diseases. This model, made publicly available, achieved an impressive accuracy of 0.96 for melanoma, 0.83 for squamous cell carcinoma, and 0.96 for basal cell carcinoma [3]. In a different study, a two-layer CNN trained on 136 images for distinguishing melanoma from benign nevi achieved 81% accuracy, though the small dataset size suggests the need for cautious interpretation of the results [4]. The incidence of melanoma, particularly among young adults, has been on the rise. In the U.S., 2,400 melanoma cases were reported among individuals aged 15 to 19.
While melanoma cases in people over 50 have increased by more than 2% annually, the rate has decreased by more than 1% each year for those under 50 [5]. To address this growing concern, data augmentation techniques have been employed to enhance CNN performance in melanoma classification. One study achieved an AUC of 89.2%, AP of 73.9%, and PPV of 82.3% on the ISIC 2017 dataset, demonstrating the positive impact of image deformation on classifier performance [6].
Transfer learning has also been effectively applied to the HAM10000 dataset, improving the classification accuracy of skin cancer lesions. The ResNet model, pre-trained on the ImageNet dataset, proved particularly effective, while traditional algorithms like Random Forest and SVM showed less promise [7]. Additionally, a deep learning pipeline based on morphological analysis of skin lesions demonstrated strong performance in early skin cancer diagnosis, further validating the effectiveness of such approaches [8].
In the realm of image segmentation, the U-Net CNN algorithm was used to segment images, followed by feature extraction methods like Edge Histogram (EH), Local Binary Pattern (LBP), and Histogram of Oriented Gradients (HOG). These features were then classified using SVM, KNN, Naïve Bayes (NB), and Random Forest (RF) classifiers on a dataset of 900 images from ISIC, with a 90% training and 10% testing split [9]. Another study modified the AlexNet model for classifying 10 types of skin lesions, achieving an accuracy of 81.8% on a dataset of 1,300 clinical images from the DermoFit Image Library [10].
Furthermore, a method using vector-based SURF with a multi-SVM classifier achieved 86.37% accuracy, 86.53% sensitivity, and 96.42% specificity on 611 images of four skin lesion types [11]. Hybrid models combining pre-trained ResNet-18, AlexNet, and VGG16 with SVMs achieved 83.83% accuracy for melanoma and 97.55% for seborrheic keratosis on 150 images from the ISIC 2017 dataset [12]. Another study utilizing DenseNet201, ResNet152, and Inception V4 models on 10,015 images achieved high confusion matrix scores, particularly for Melanocytic nevus, with a 2% improvement observed when using DenseNet201 by cropping images during training [13].
CNNs trained on datasets like ImageNet and Dermnet have also shown promising results, with one study using AlexNet achieving an accuracy of 89.3%, sensitivity of 77.1%, and specificity of 93.0% in classifying skin diseases [14]. Additionally, a new CNN-based prediction model using a novel regularizer achieved 97.49% accuracy in determining whether lesions were benign or malignant on 8,000 images from ISIC, with AUC scores ranging from 0.77 to 0.93 across different lesion comparisons [15]. Addressing the challenges of small datasets, another study proposed using sub-images as input to a CNN with Fisher vector encoding and SVM classifiers, achieving 83.09% accuracy on 1,279 images from the ISBI 2016 dataset [16].
Despite the advancements in CNN-based methods, traditional models such as the ABCD rules, the seven-point checklist, and various mobile applications for skin cancer detection still face limitations in performance and efficiency, especially when restricted to specific image types like dermoscopy or histopathology. These conventional methods, while helpful, often fall short in accuracy and reliability compared to the newer, more sophisticated deep learning approaches.
B. Issues with the existing solutions
Convolutional Neural Networks (CNNs) have consistently demonstrated superior performance in skin lesion classification compared to other architectures. Notably, models like AlexNet, which won the Large-Scale Visual Recognition Competition (ILSVRC) in 2012, and ResNet, the winner in 2015, have been widely used in skin cancer detection. Studies using AlexNet for skin classification reported accuracies ranging from 81.8% for 150 images to 90.3% for 1,300 images of 10 skin lesion types. However, significant variability in results was observed, particularly when comparing studies using different datasets and numbers of skin lesion types. For instance, one study using 8 lesion types showed different outcomes compared to another using 12 types, highlighting the challenges in standardizing results across various research efforts.
A critical issue in current research is the underrepresentation of dark-skinned individuals in skin lesion datasets, which are predominantly sourced from light-skinned populations in the United States, Australia, and Europe. To improve classification accuracy across diverse skin tones, it is essential to train CNNs with a more representative dataset that includes images from dark-skinned people. Additionally, incorporating clinical data such as age, gender, and skin type could further enhance the performance of these classifiers.
In reviewing multiple studies, it became apparent that comparing results is often challenging due to differences in methodologies and the amount of data used. To address this, future research should utilize publicly available benchmarks and fully disclose their methods to facilitate comparability. Moreover, the potential impact of adversarial attacks and the need for balanced datasets, including both positive and negative examples, should be considered to avoid biases and improve the robustness of CNN-based skin lesion classification models. Future publications should prioritize these aspects to advance the accuracy and reliability of CNNs in dermatological applications.
III. OVERVIEW OF WORK
A. Problem Definition
In this project, we aim to develop a deep learning model using a Convolutional Neural Network (CNN) to predict new cases of skin cancer. The first phase involves preparing image data, including segmentation to identify regions of interest for more effective analysis. Our proposed CNN model comprises three convolutional layers, three max-pooling layers, and four fully connected layers. Testing this model has yielded promising results, achieving an accuracy of 90%.
Additionally, we plan to build a web application that utilizes this model to provide online diagnosis of skin cancer lesions. The primary objective of this project is to create a state-of-the-art CNN model capable of classifying skin lesion images into various cancer types. The model is trained and tested on datasets provided by the International Skin Imaging Collaboration (ISIC) and MNIST HAM, enabling early detection of potentially dangerous lesions.
B. Proposed System and Methodology
After extensive research, we propose a new, efficient model for accurately classifying and detecting skin cancer types without the need for clinical procedures, using a fine-tuned MobileNet Convolutional Neural Network (CNN). This model classifies skin cancer lesions into seven distinct classes, with all training performed within a specialized kernel.
Notably, much of the current CNN research has focused on skin lesions from light-skinned individuals, primarily from regions like Eastern Europe and North America. To address this bias, it is crucial to train CNNs to account for diverse skin tones, ensuring accurate classification across different skin colors. Our model also seeks to improve classification quality by incorporating clinical data such as age, image size, gender, and skin type as additional inputs for the classifiers.
Moreover, we leverage TensorFlow.js, a new library that enables machine learning models to run directly in the browser, ensuring user data remains local without requiring any downloads or installations. The primary goal of our model is to facilitate early detection of skin cancer, enabling effective treatment and reducing mortality rates. We will also compare our model's performance with other datasets to demonstrate its superiority.
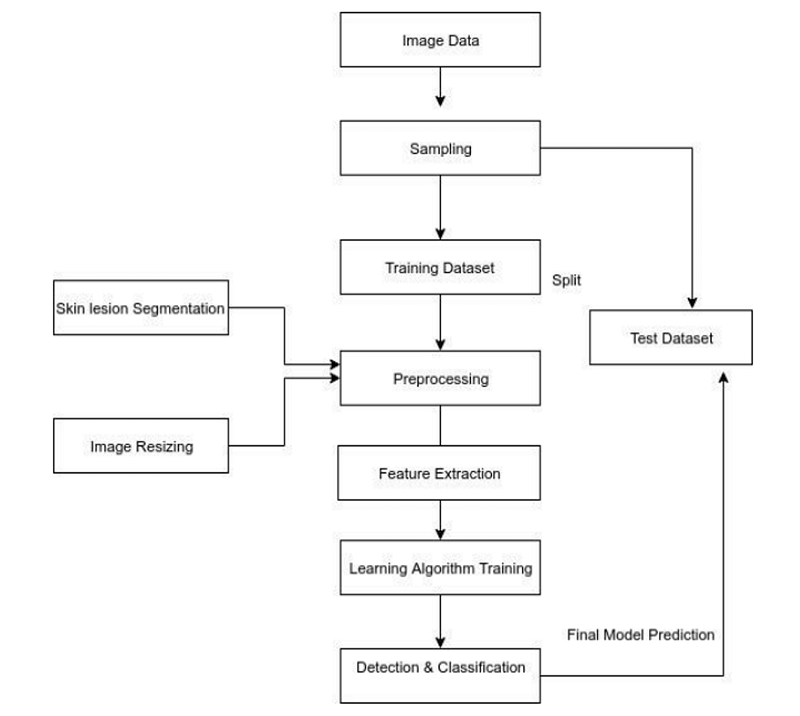
Fig. 1 Methodology
IV. SYSTEM DESIGN
A. Algorithm
The basic pipeline for CNN is as follows:
- Input an image.
- Perform Convolution operation to get an activation map.
- Apply the pooling layer to make our model robust.
- Activation function (mostly ReLu) is applied to avoid non-linearity.
- Flatten the last output into one linear vector.
- The vector is passed to a fully connected artificial neural network.
- The fully connected layer will provide a probability for each class that we’re after.
- Repeat the process to get well defined trained weights and feature detectors.
B. Convolution layer
A convolution filter extracts features from an image by learning patterns as it processes the image. It does this by computing a dot product between the filter values and the image pixel values, forming a convolution layer. The values within the filter matrix are continuously updated during the backpropagation process to improve accuracy. However, the dimensions of the filter matrix are predefined by the programmer, allowing control over the scale of features being detected.
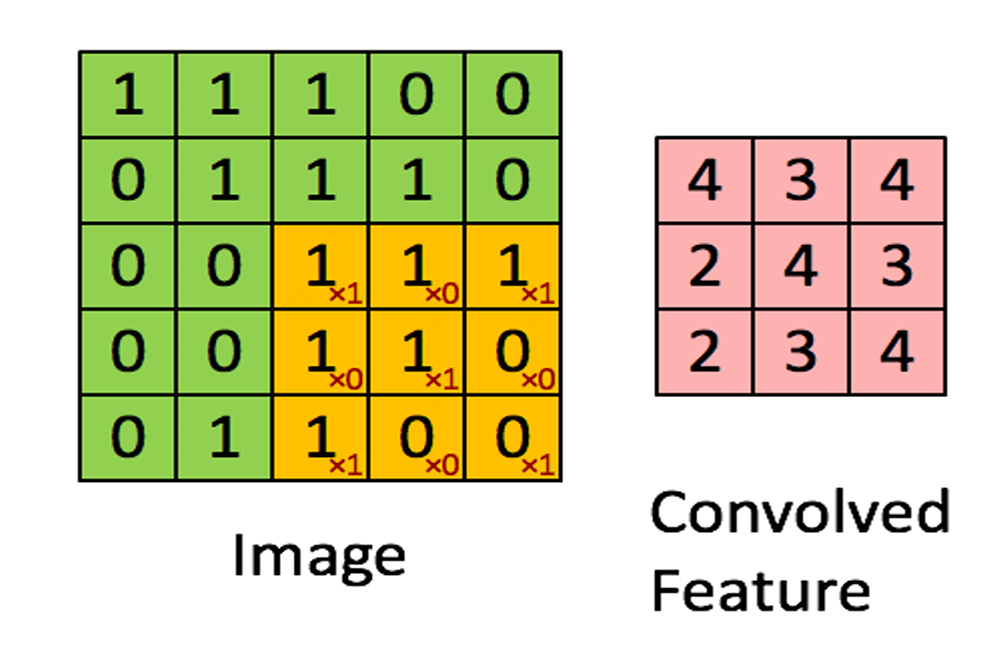
Fig. 2 Convoluting a 5x5x1 image with a 3x3x1 kernel to get a 3x3x1 convolved feature
When applying convolution filters to a 3D input, each filter produces a 2D output. By combining these 2D outputs, a final 3D output is obtained. The dimensions of this output are reduced from (h, w, c) to (h-f+1, w-f+1, c) because the convolution operation cannot be applied at the boundaries without crossing the input’s edges.
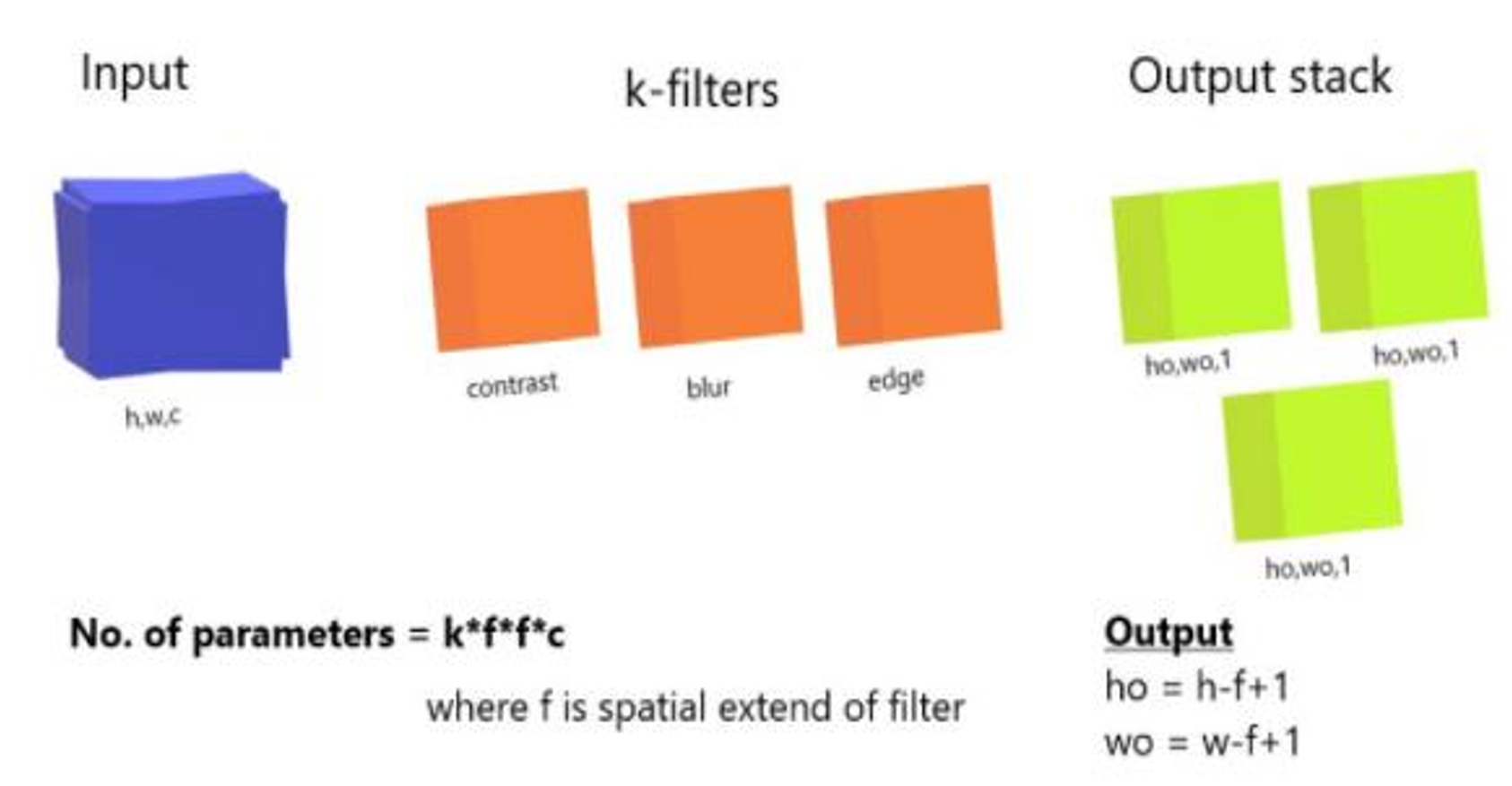
Fig. 3 3D filtering
To retain the original dimensions of the output image, padding is used by adding zeros around the edges of each channel.
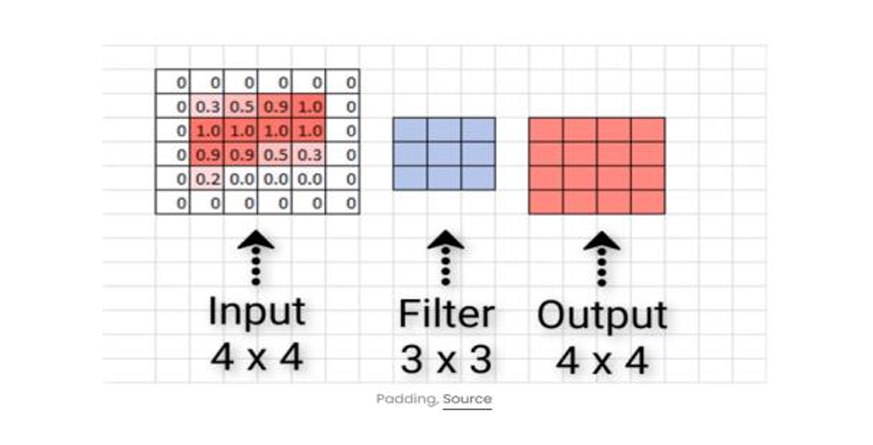
Fig. 4 Zero Padding
In convolution, stride allows us to control how many pixels we skip while moving the filter across the input, either vertically or horizontally. This results in a more customized activation map, compared to the usual one-pixel skip.
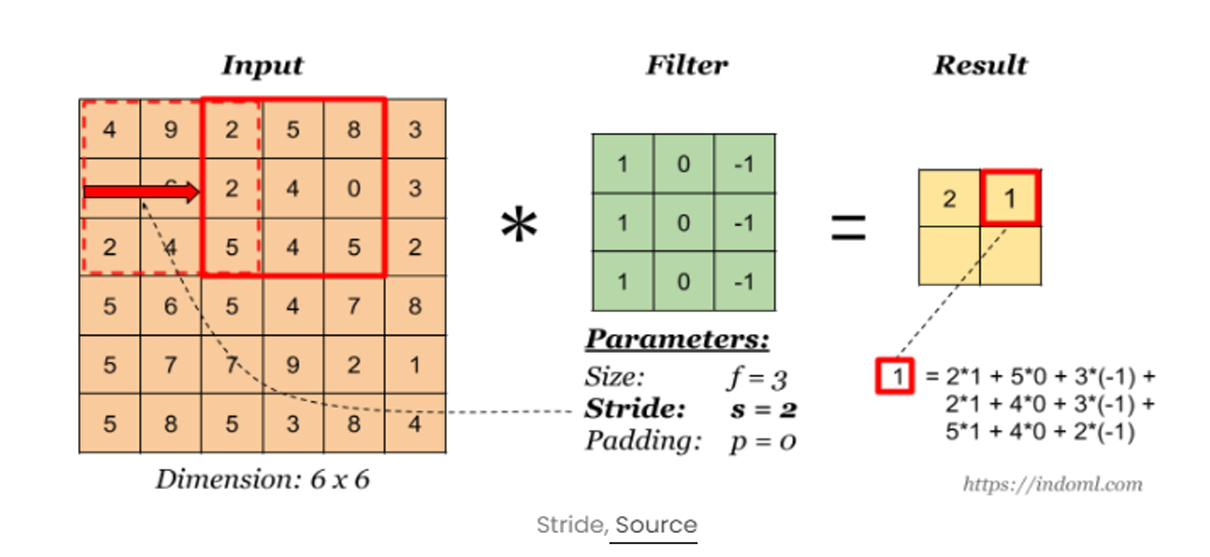 Fig. 5 Convolutional operation
Fig. 5 Convolutional operation
2) Pooling layer
CNNs enhance model robustness and feature detection by replacing the output with a max summary, reducing data size and processing time. This approach helps highlight the most significant features.
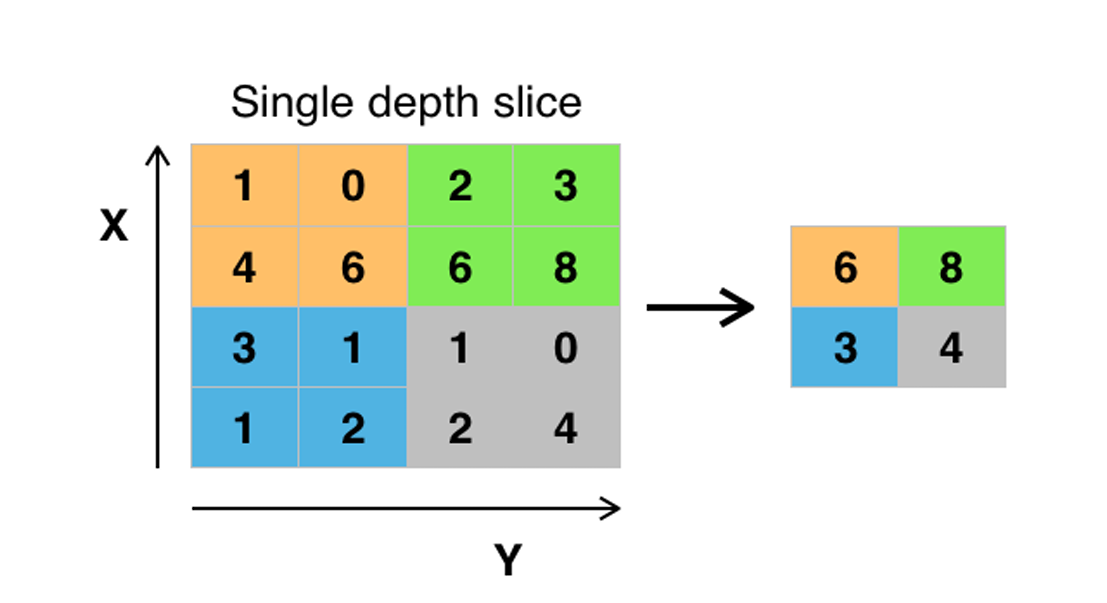 Fig. 6 Pooling layer
Fig. 6 Pooling layer
Max pooling takes two hyperparameters: stride and size. The stride determines how many values are skipped, while the size defines the area over which each maximum value is selected.
3) Activation Function (ReLU and Sigmoid):
After applying convolution and pooling operations, activation functions like ReLU are introduced to add nonlinearity. ReLU outputs the input directly if it's positive (x > 0) and returns 0 otherwise, effectively addressing diminishing gradient issues. Small weights are set to 0 after ReLU activation.
4) Fully Connected Layer:
At this stage, we add an artificial neural network as the final layer of our convolutional neural network. The flattened feature output is transformed into a column vector and passed through the SoftMax activation function, which assigns probabilities to each class.
Each node in the previous layer connects to the final layer, representing the distinct labels. Finally, backpropagation is used to iteratively adjust weights and biases, optimizing the model's performance through continuous learning.
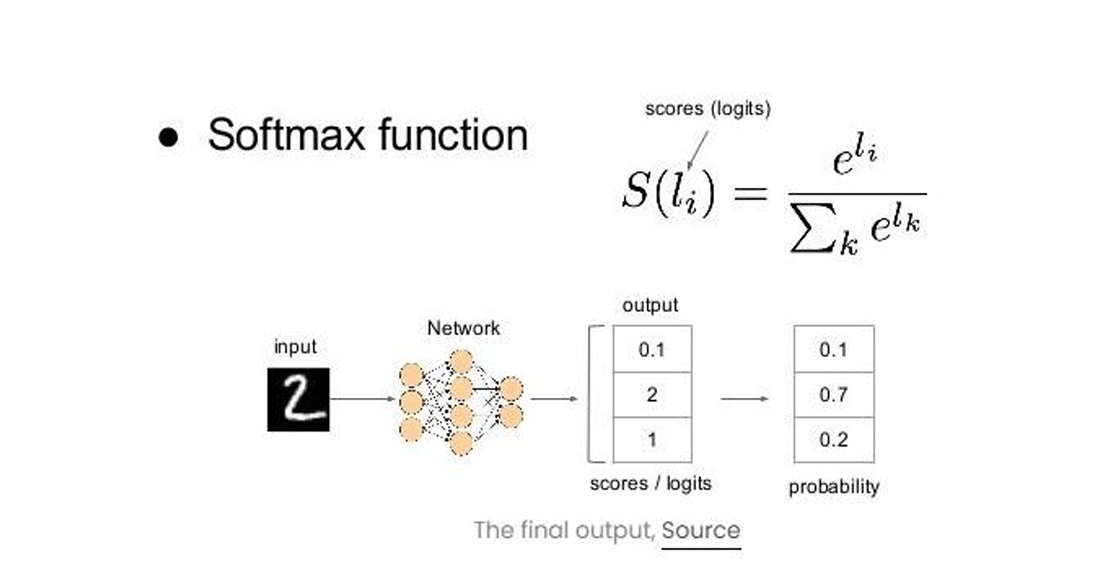 Fig. 7 Softmax function
Fig. 7 Softmax function
In a model with two output classes—cat and dog—specific neurons are triggered by distinct features. For instance, cat neurons activate when features like "whiskers," "cat-iris," and "small, light body" are identified, while dog neurons respond to "big wet nose," "floppy ears," and "heavy body." Through iterative training, the model learns to distinguish and prioritize these key features, assigning greater weight to them, which improves its ability to accurately classify images as either a cat or a dog.
B. Architecture
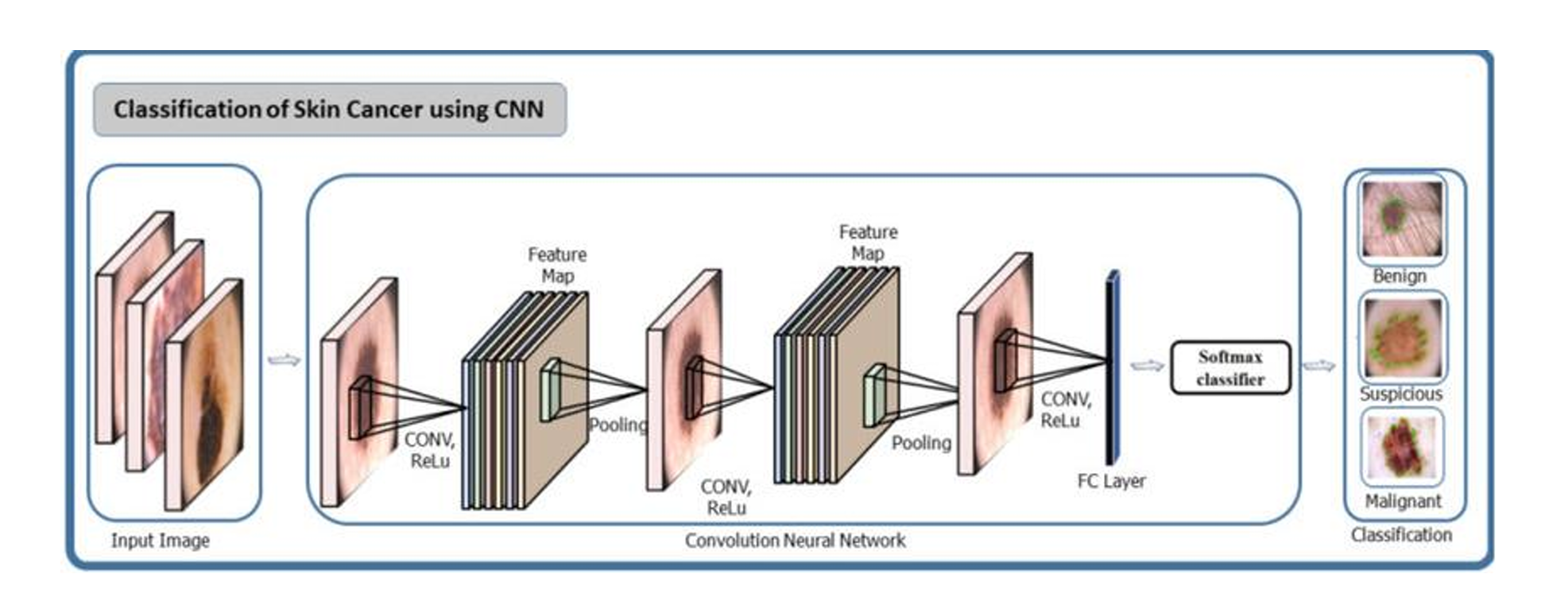
Fig. 8 Architecture diagram
C. Block diagram
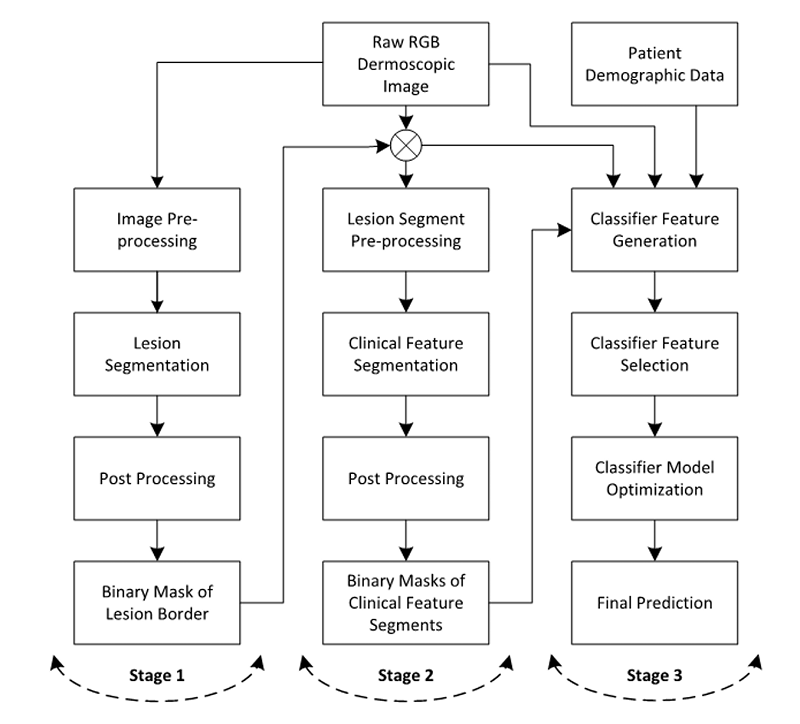
Fig. 9 Block diagram of computerised detection of melanoma
D. Dataset
The dataset we’ve used to propose our model was taken from Kaggle, entitled “Skin Cancer MNIST: KAM10000”. It contains a large collection (read 10015) of multi-source dermatoscopic images of all important diagnostic categories in the realm of pigmented lesions. It was be used to compare the accuracies between different stages of melanoma.
The results coming out of it has been compared with three other datasets (2016, 2017, 2018 versions) of another league, taken from the archives of ISIC.
V. IMPLEMENTATION AND OUTPUT
A. Training the model
We’re training and validating the dataset(s) into a MobileNet CNN model which further will be used in the deployment of a web app.
B. Web deployment
We’re using Tensorflow.js for converting the Keras model.h5 to a more compact form under model.json, which will predict the images uploaded by the user in the backend.
C. Test cases
If any file formats except .png, .jpg, .jpeg, .jfif are uploaded, no significant result will be displayed.
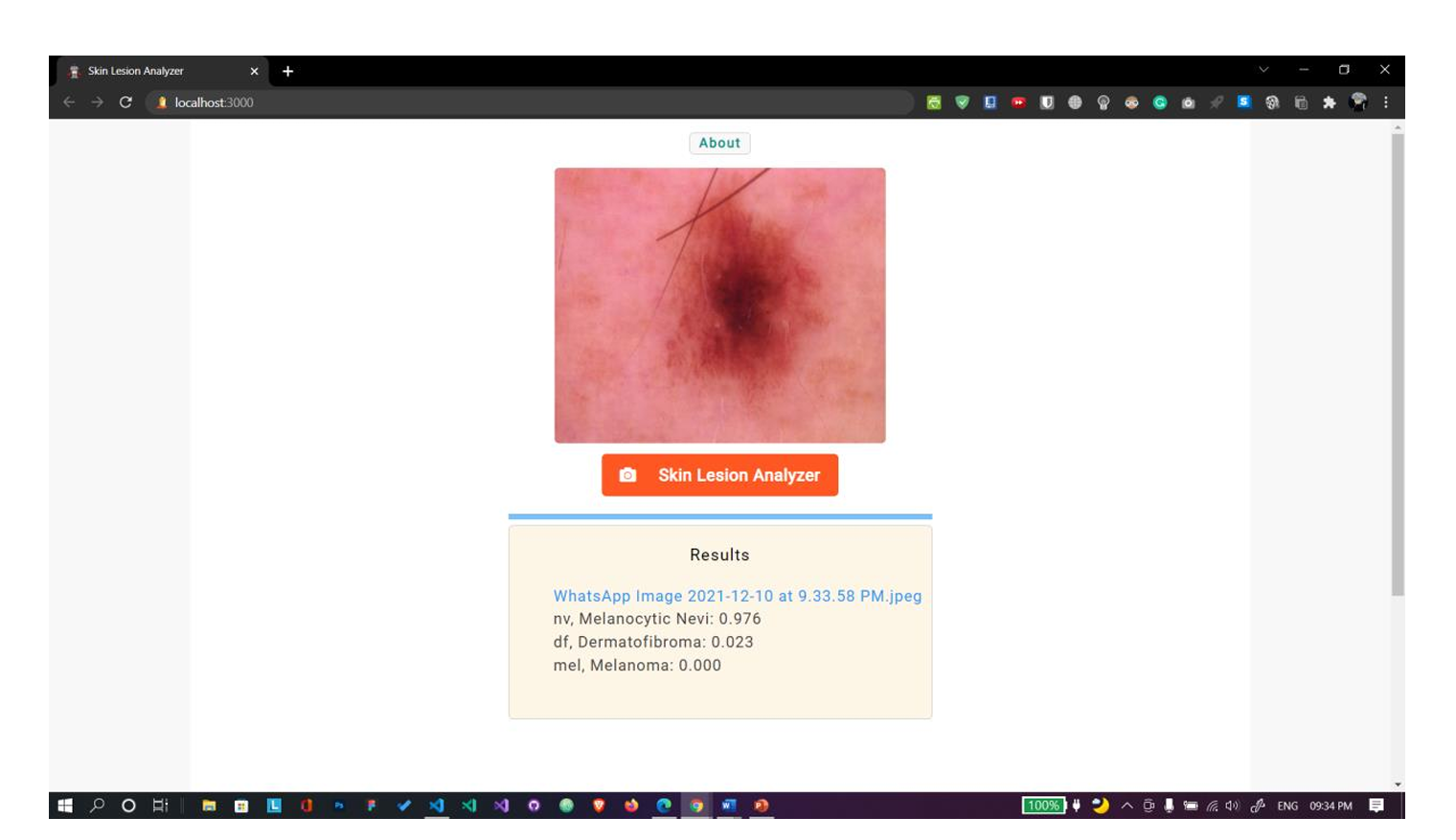
Fig. 10 Uploaded file is in jpeg format. Shows correct results
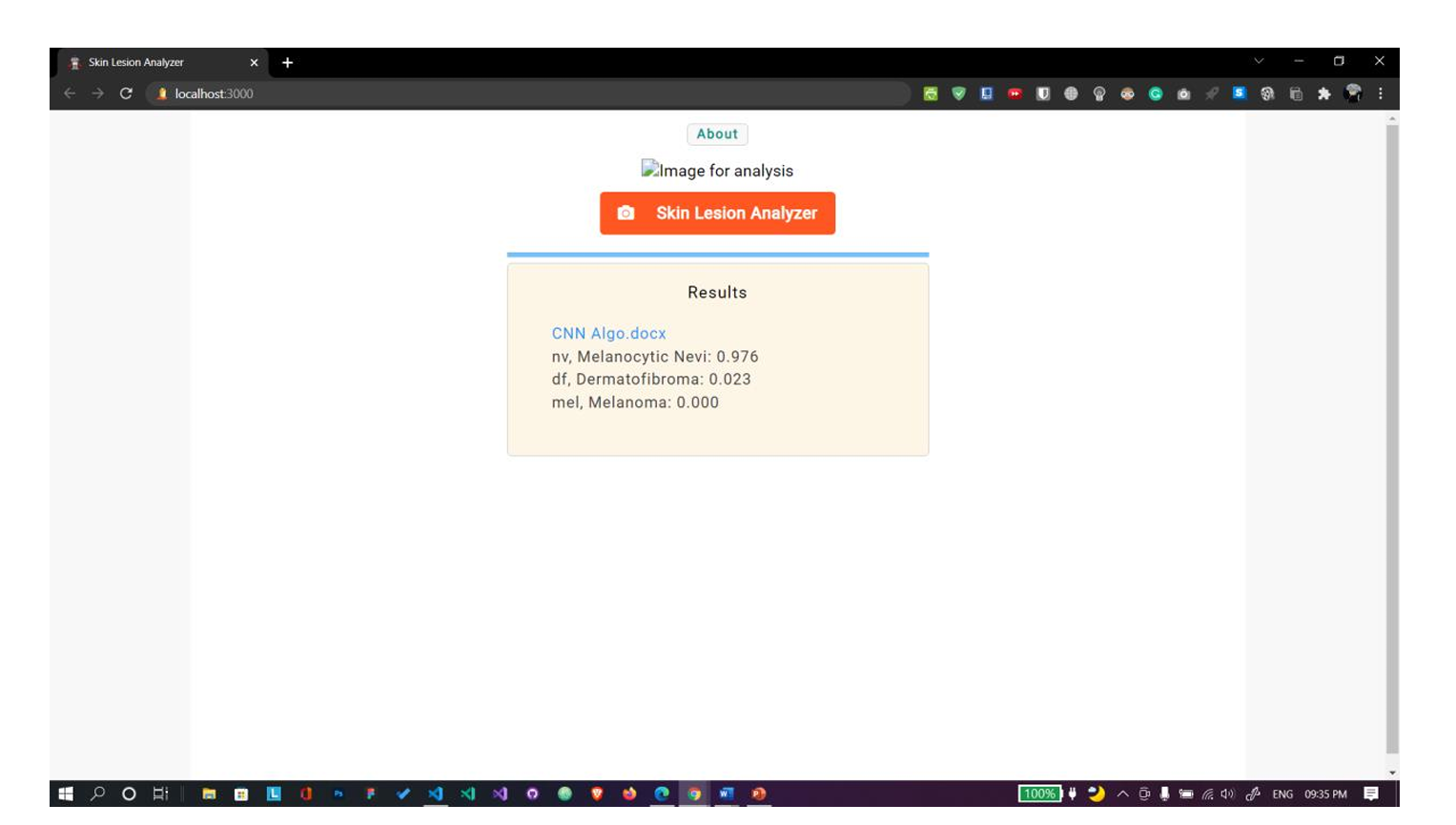 Fig. 11 Uploaded file is in docx format. Shows no results.
Fig. 11 Uploaded file is in docx format. Shows no results.
D. Execution snapshots
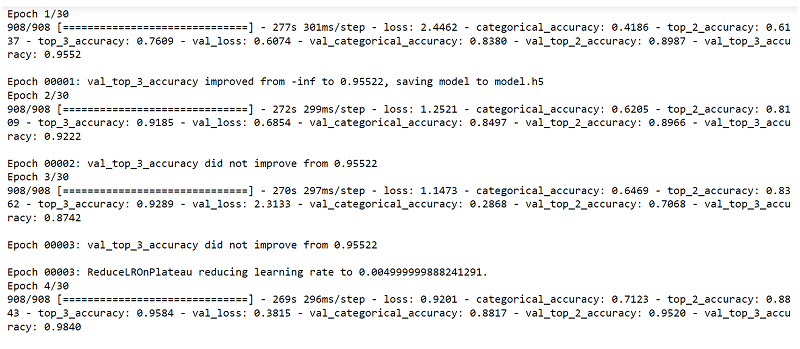
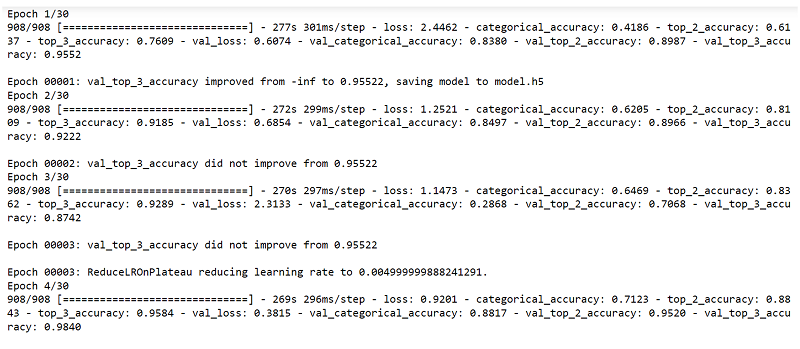
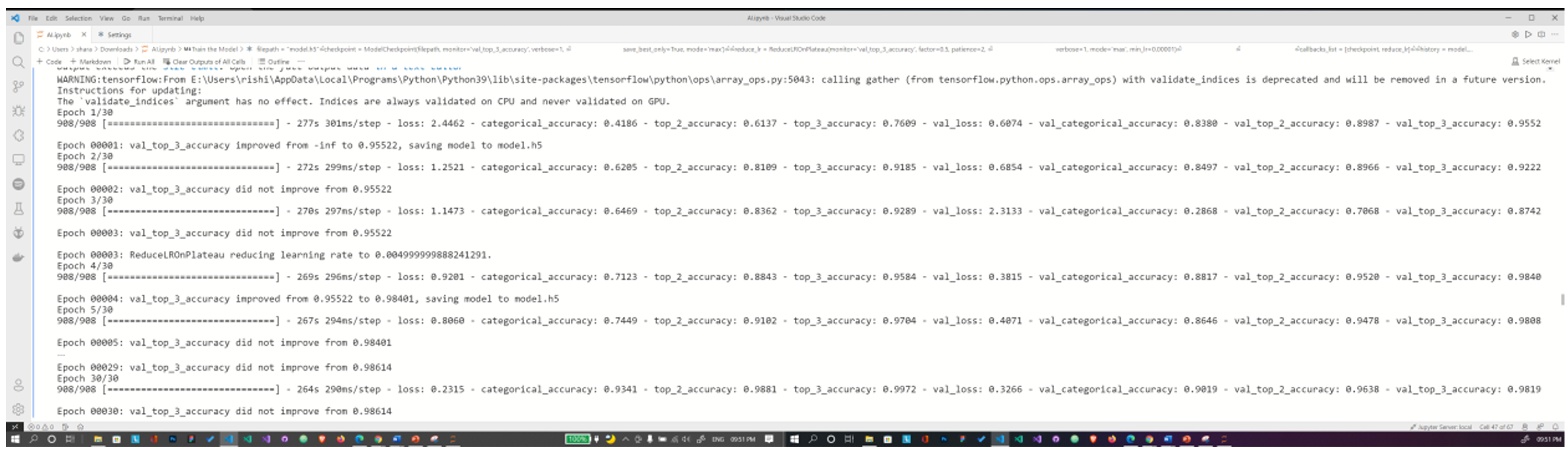 Fig. 12 Training the model
Fig. 12 Training the model

E. Skin lesion analyzer
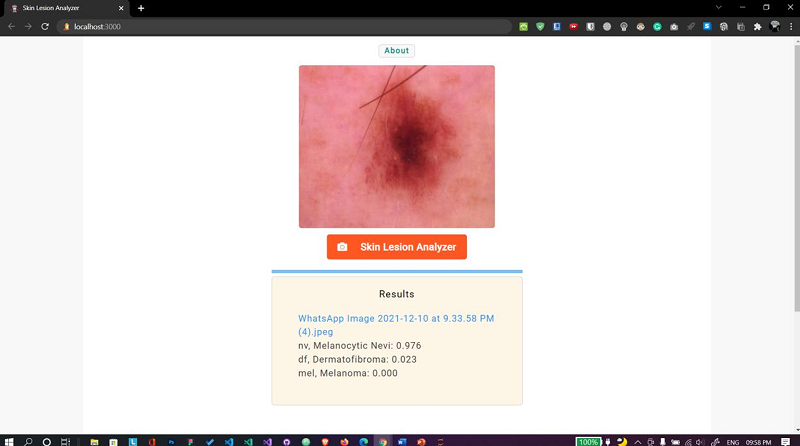
Fig. 13 Web app Case 1
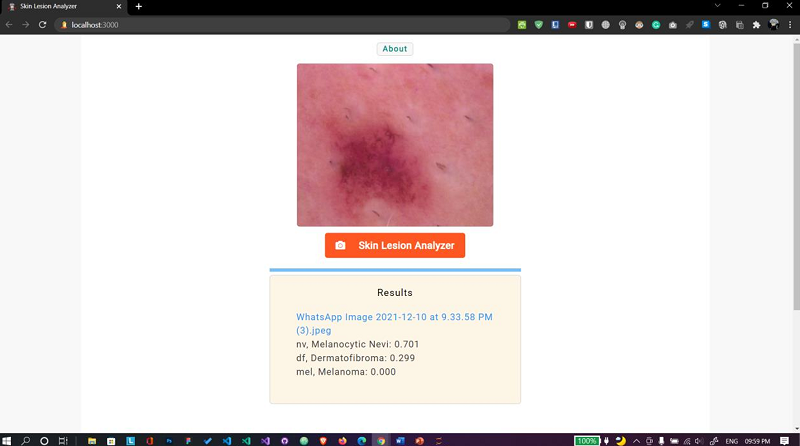
Fig. 14 Web app Case 2
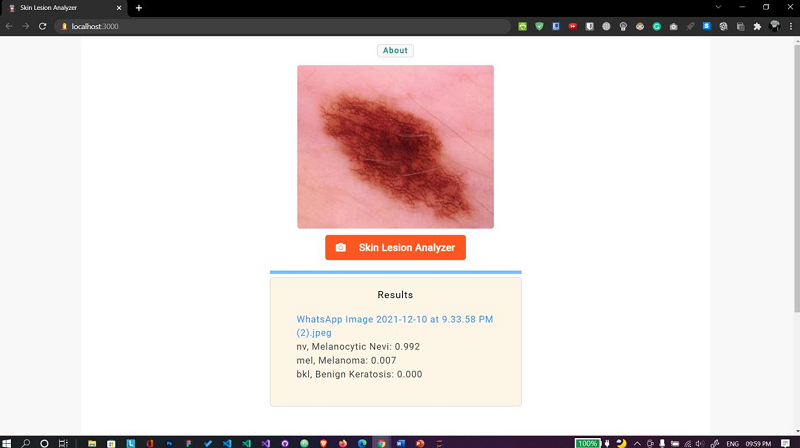
Fig. 15 Web app Case 3
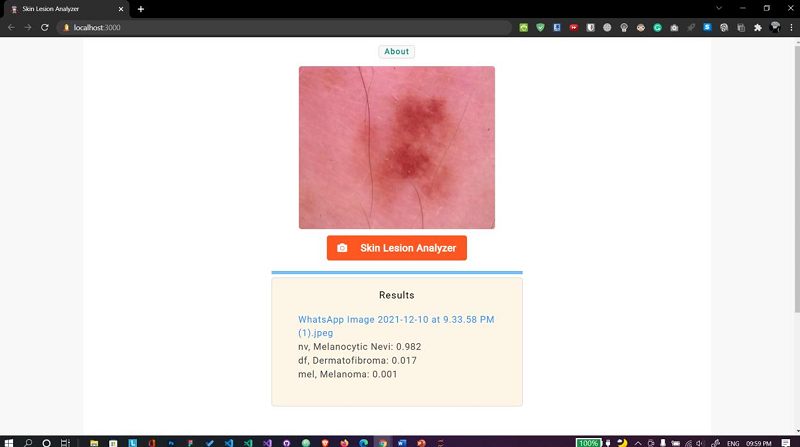
Fig. 15 Web app Case 4
F. Output
1) Performance Metrics:
TABLE I
Performance Metrics Comparison
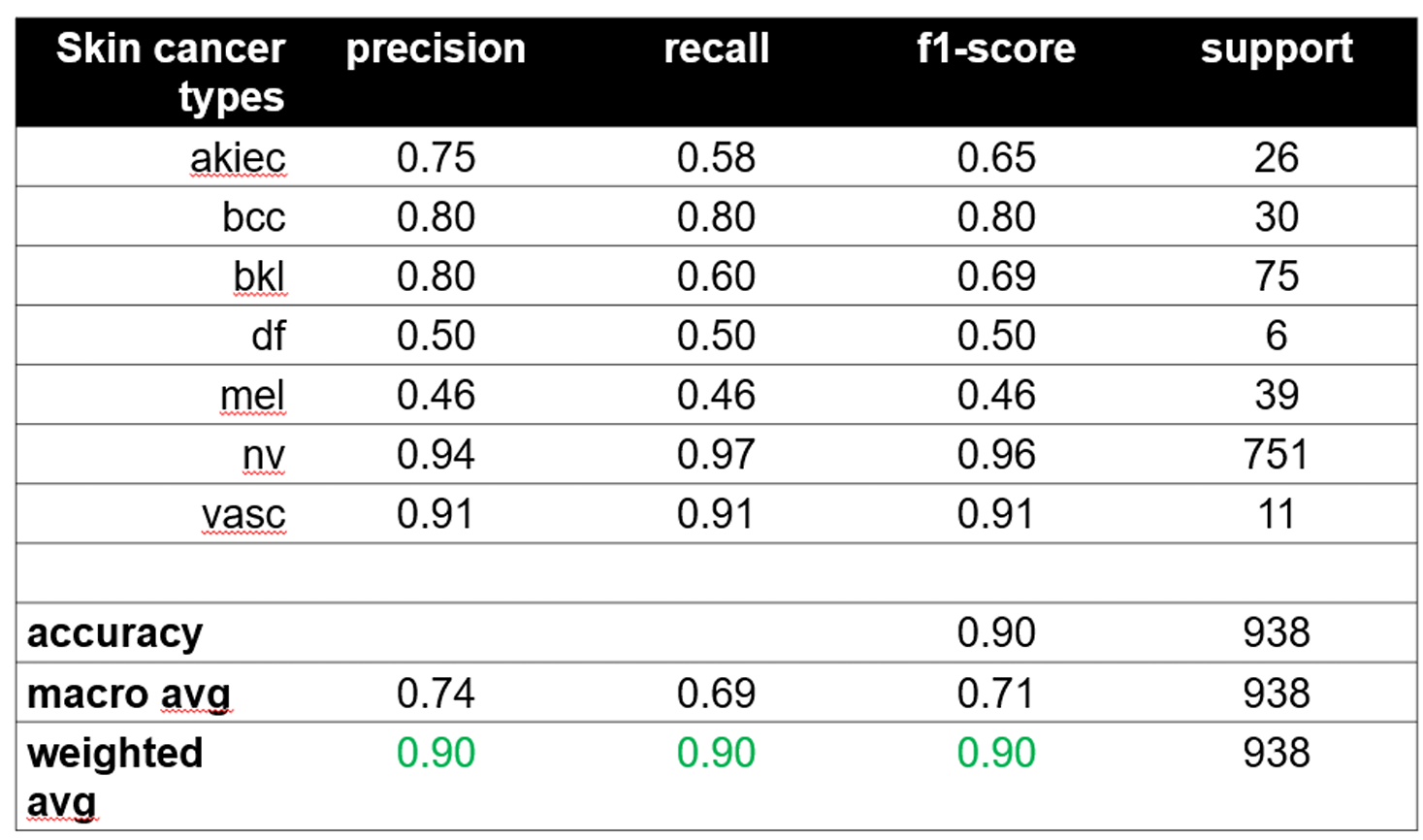
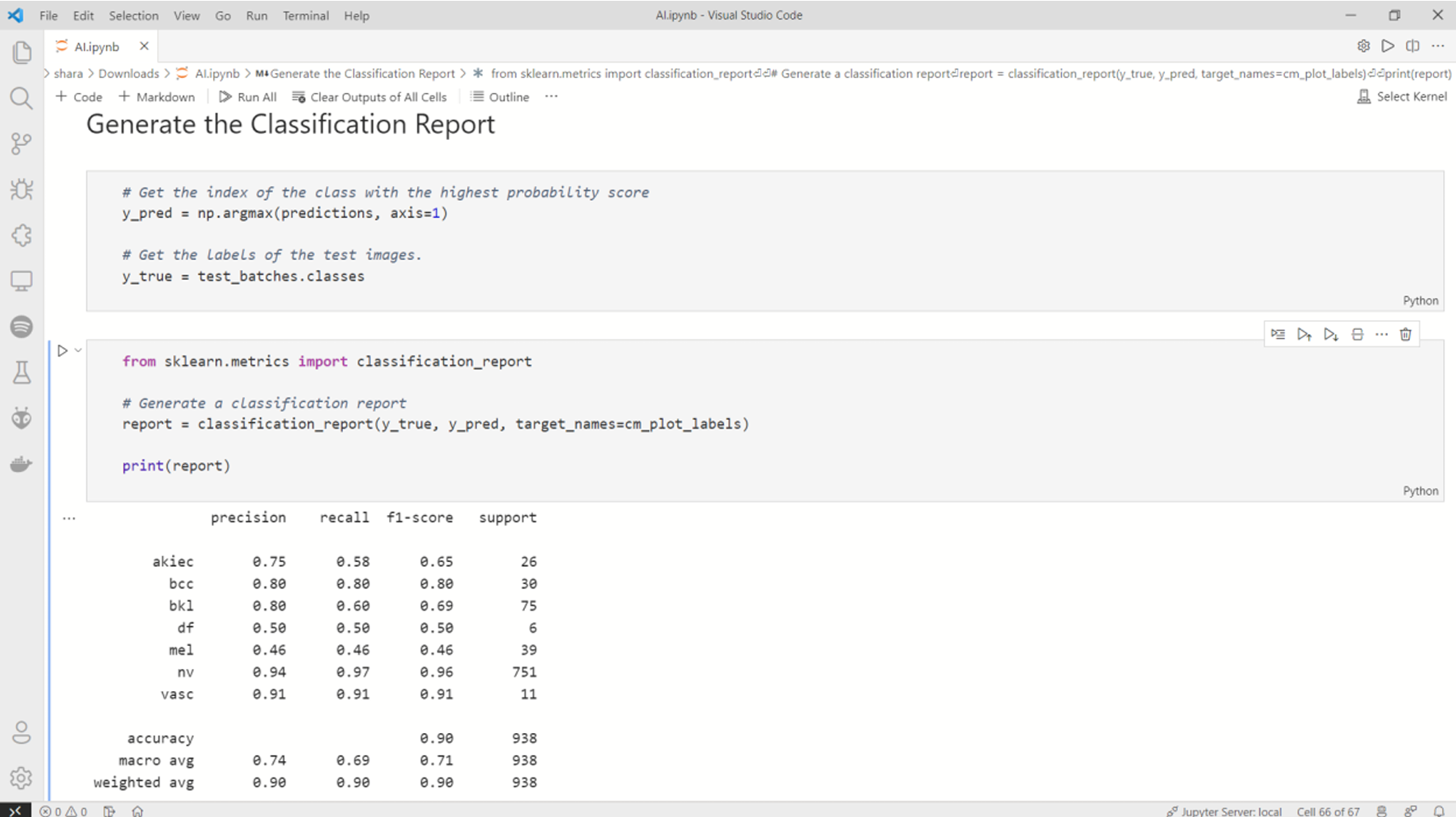 Fig. 16 Performance metrics
Fig. 16 Performance metrics
2) Confusion Matrix:
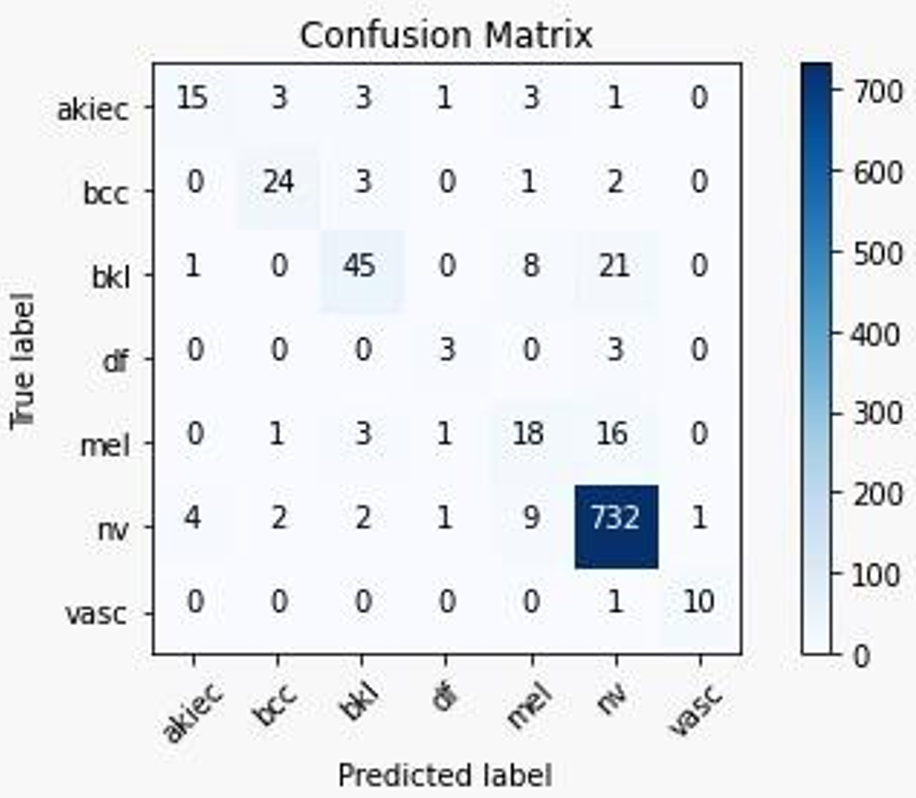
 Fig. 17 Confusion Metrix
Fig. 17 Confusion Metrix
3) Training curves
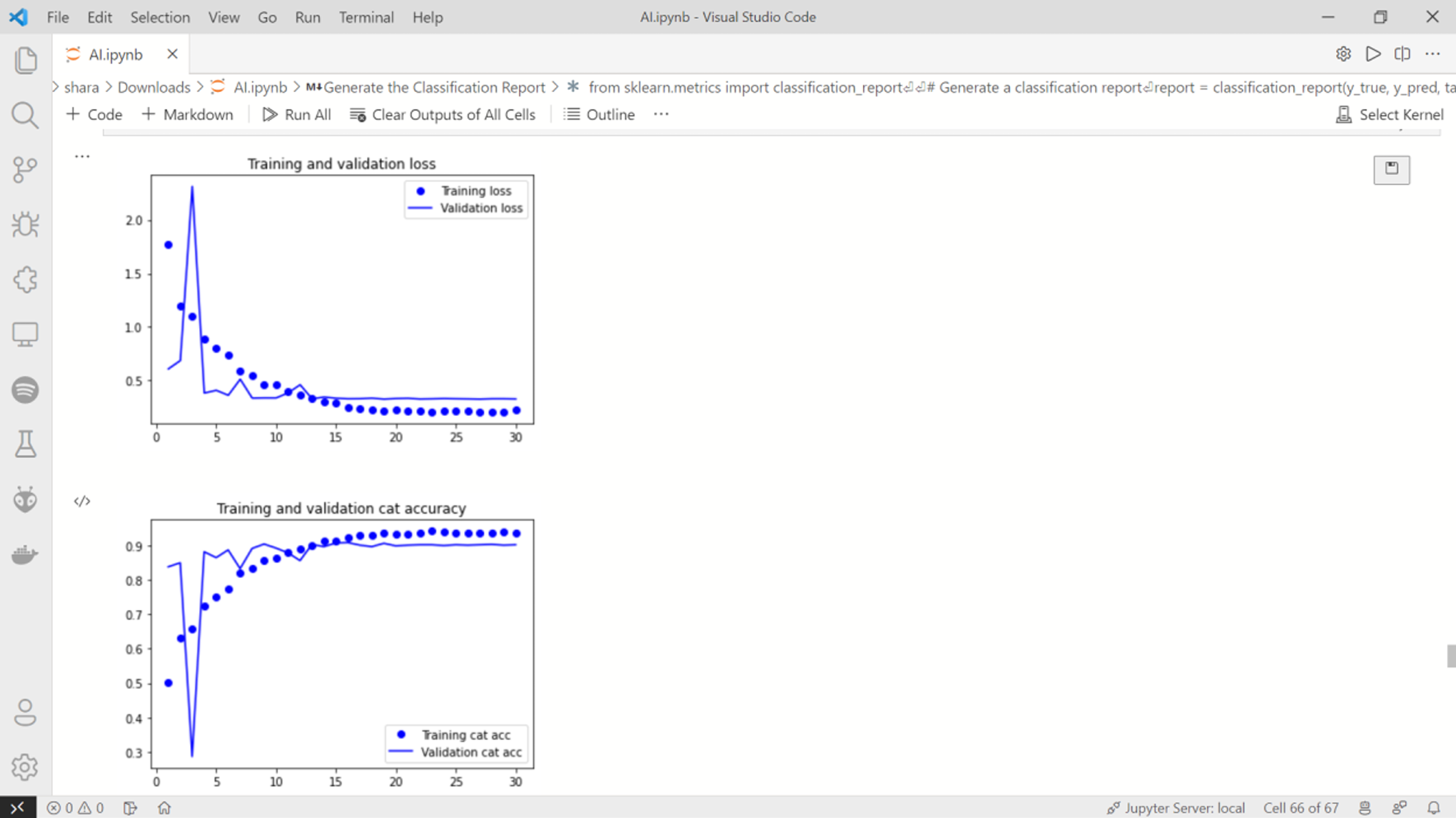 Fig. 18 Training curves 1 & 2
Fig. 18 Training curves 1 & 2
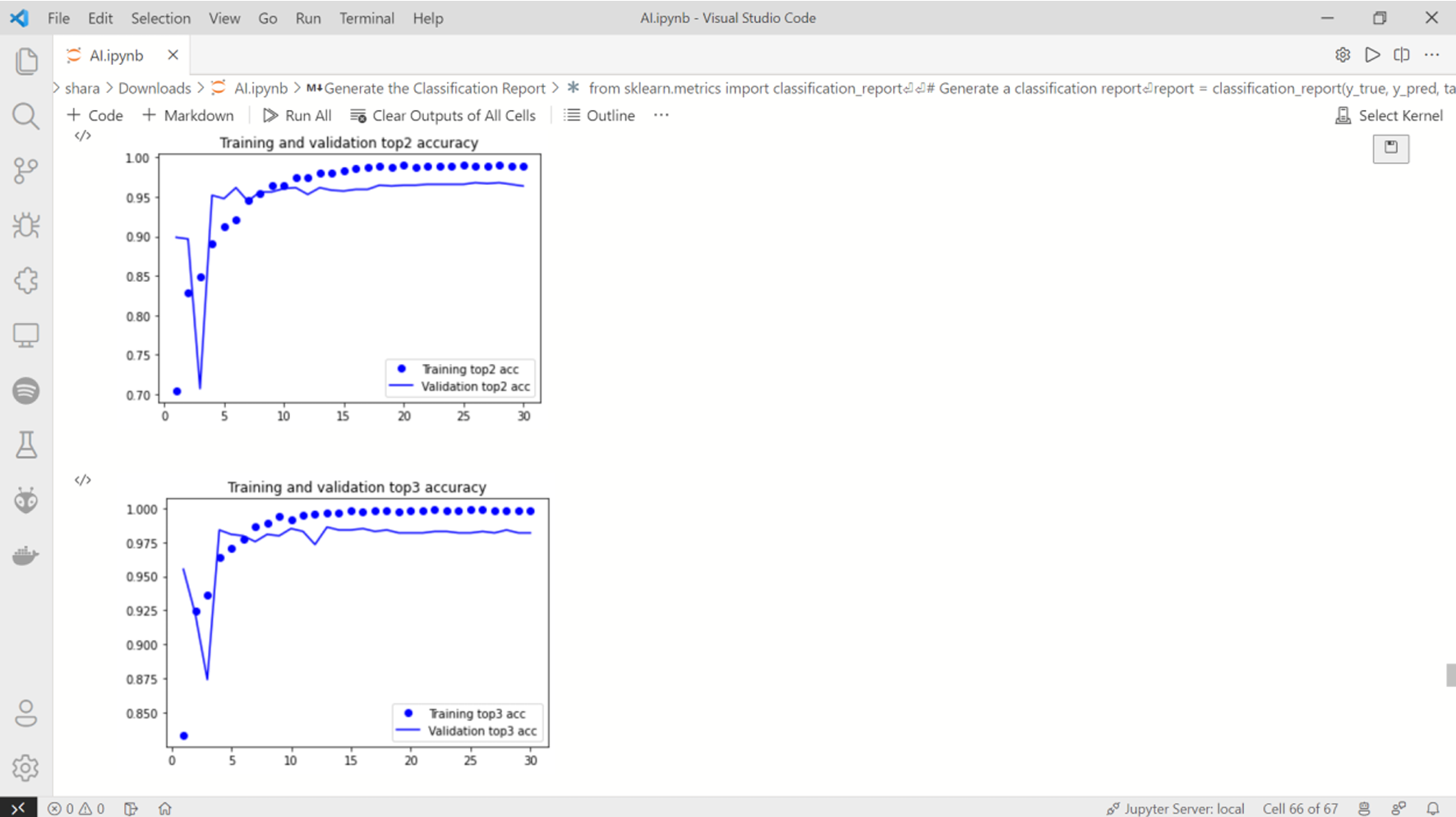 Fig. 19 Training curves 3 & 4
Fig. 19 Training curves 3 & 4
G. Performance Comparison
TABLE 2
Performance Comparison of various Datasets
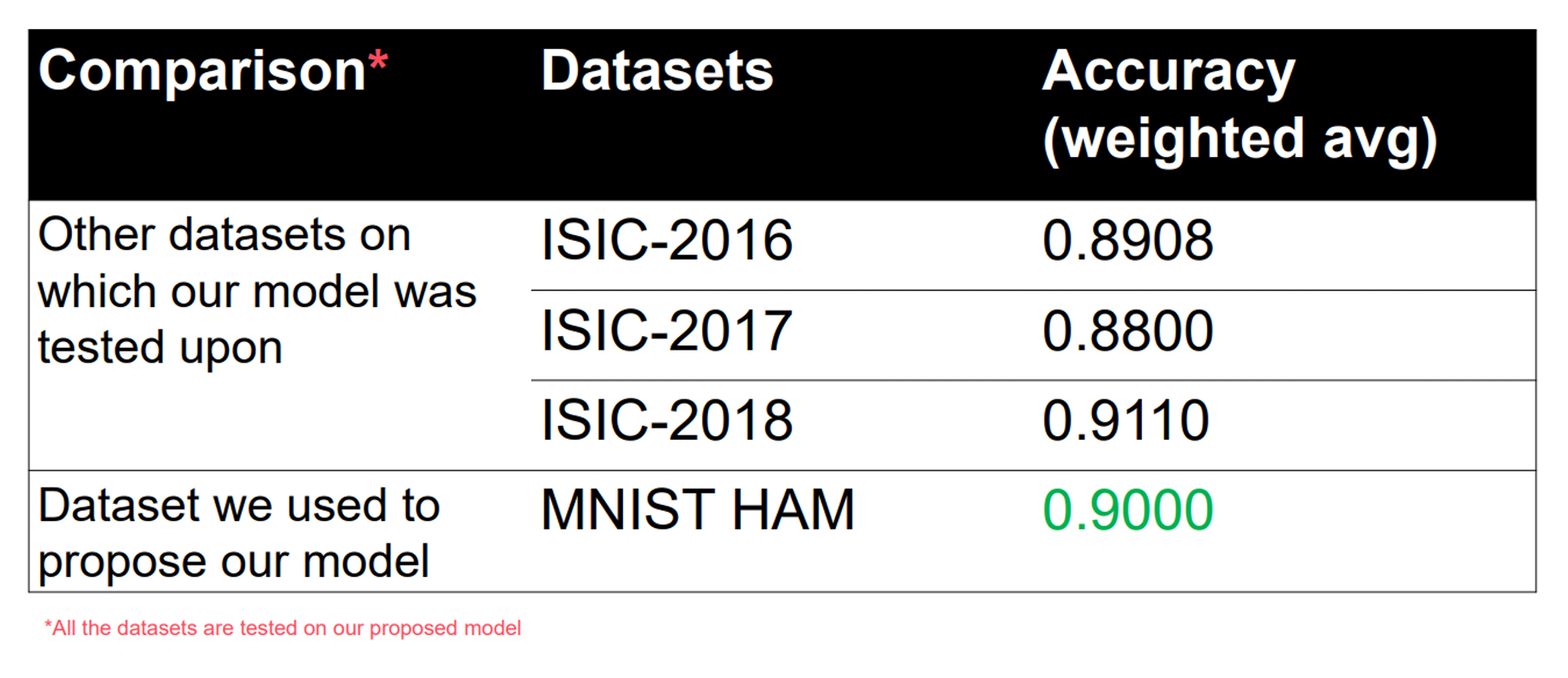
Using matplotlib library, we’ve compared the difference in results and plotted them in a bar graph.
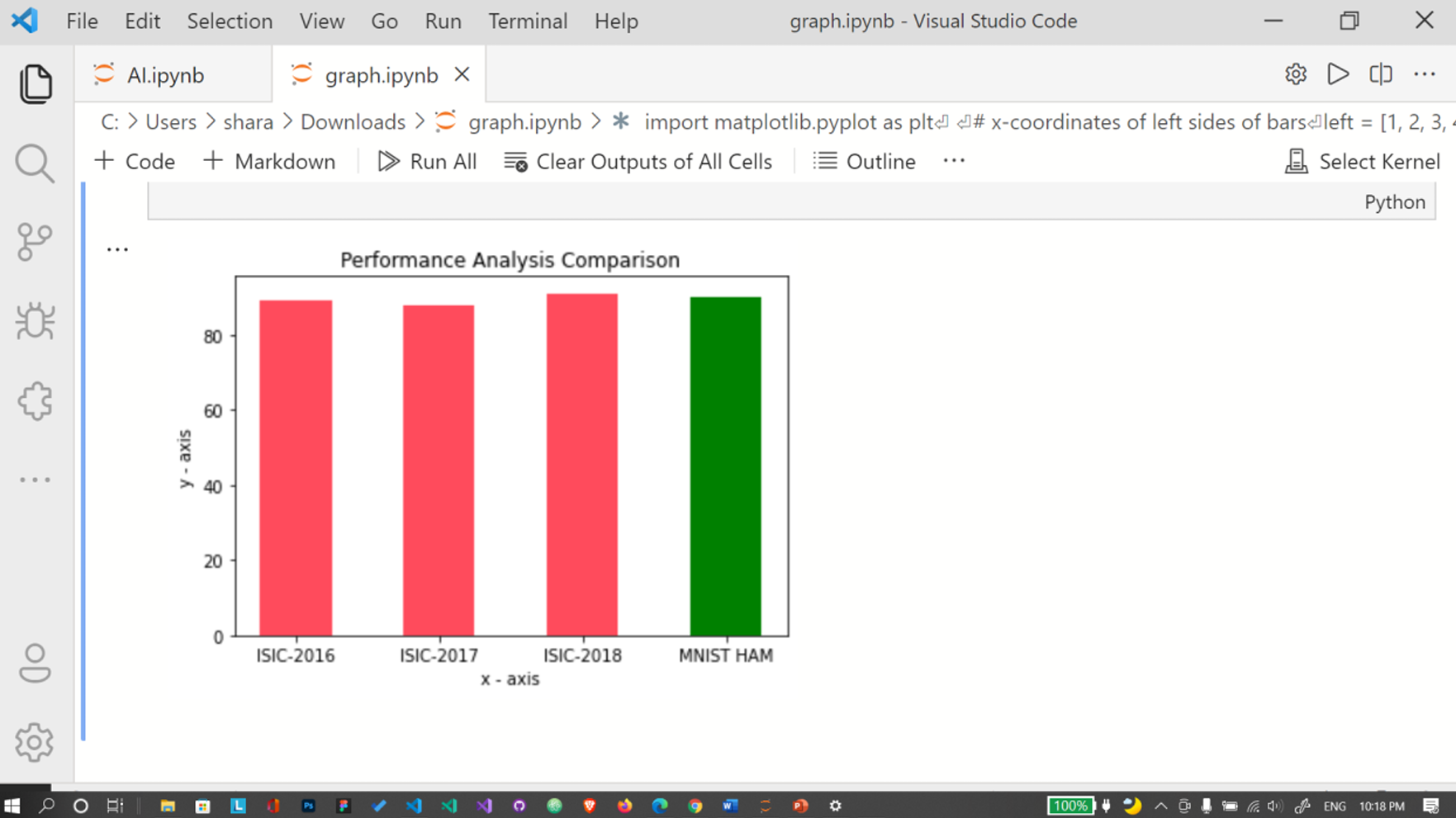 Fig. 20 Performance Analysis Comparison Graph
Fig. 20 Performance Analysis Comparison Graph
Conclusion
This project aimed to develop a convolutional neural network (CNN) model for diagnosing and detecting skin cancer from lesion images. It also investigated the use of data augmentation as a preprocessing technique to enhance the classification robustness of the CNN model. Our approach involved an initial step of feature extraction, followed by training and testing the model using transfer learning. Based on our observations, we concluded that the transfer learning technique can be effectively applied to the HAM10000 dataset to improve the classification accuracy of skin cancer lesions. Additionally, we found that the ResNet model, pre-trained on the ImageNet dataset, significantly enhances the accurate classification of cancerous lesions in the HAM10000 dataset. Furthermore, we compared the results obtained from the HAM10000 dataset with those from the ISIC datasets. In most cases, the HAM10000 dataset yielded better performance, consistently achieving accuracy rates of 90% or higher. Building on these promising results, future work will focus on further improving prediction outcomes and classification accuracy. This may involve integrating personalized patient data, such as genetic information, age, and skin tone, into the current study for skin cancer diagnosis. Incorporating these additional features could be beneficial in developing personalized computer-aided systems for diagnosing skin cancers.
References
[1] N. Zhang, Y.-X. Cai, Y.-Y. Wang, Y.-T. Tian, X.-L. Wang, and B. Badami, \"Skin cancer diagnosis based on optimized convolutional neural network,\" Artif. Intell. Med., vol. 102, 2020. [2] L. Zhang, H. J. Gao, J. Zhang, and B. Badami, \"Optimization of the Convolutional Neural Networks for Automatic Detection of Skin Cancer,\" Open Med., vol. 15, no. 1, pp. 27-37, 2020. [3] M. Karthiga, R. K. Priyadarshini, and A. B. Banu, \"Malevolent Melanoma diagnosis using Deep Convolution Neural Network,\" Res. J. Pharm. Technol., vol. 13, no. 3, pp. 1248-1252, 2020. [4] M. Chen, W. Chen, W. Chen, L. Cai, and G. Chai, \"Skin Cancer Classification with Deep Convolutional Neural Networks,\" J. Med. Imaging Health Inform., vol. 10, no. 7, pp. 1707-1713, 2020. [5] N. C. F. Codella et al., \"Skin lesion analysis toward melanoma detection,\" in Proc. Int. Symp. Biomed. Imaging (ISBI), 2017. [6] T. C. Pham, C. M. Luong, M. Visani, and V. D. Hoang, \"Deep CNN and Data Augmentation for Skin Lesion Classification,\" in Intelligent Information and Database Systems. ACIIDS 2018. Lecture Notes in Computer Science, vol. 10752, Springer, Cham, 2018, pp. 573-582. [7] R. Garg, S. Maheshwari, and A. Shukla, \"Decision Support System for Detection and Classification of Skin Cancer Using CNN,\" in Innovations in Computational Intelligence and Computer Vision. Advances in Intelligent Systems and Computing, vol. 1189, Springer, Singapore, 2021, pp. 19-27. [8] F. Rundo, G. L. Banna, and S. Conoci, \"Bio-Inspired Deep CNN Pipeline For Skin Cancer Early Diagnosis,\" Computation, vol. 7, no. 3, p. 44, 2019. [9] R. D. Seeja and S. A. Suresh, \"Deep Learning Based Skin Lesion Segmentation And Classification Of Melanoma Using Support Vector Machine (SVM),\" Asian Pac. J. Cancer Prev., vol. 20, no. 5, pp. 1555-1561, 2019. [10] J. Kawahara, A. BenTaieb, and G. Hamarneh, \"Deep features to classify skin lesions,\" in IEEE Int. Symp. Biomed. Imaging (ISBI), 2016. [11] F. Thompson and M. K. Jeyakumar, \"Vector based classification of dermoscopic images using SURF,\" Int. J. Adv. Eng. Res., 2017. [12] A. Mahbod, R. Ecker, and I. Ellinger, \"Skin lesion classification using hybrid deep neural networks,\" in Proc. IEEE Int. Symp. Biomed. Imaging (ISBI), 2017. [13] P. Tschandl, \"The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions,\" Harvard Dataverse, 2018. [14] A. Nylund, \"To be, or not to be Melanoma: Convolutional neural networks in skin lesion classification,\" Ph.D. dissertation, School Technol. Health, KTH Roy. Inst. Technol., Stockholm, Sweden, 2016. [15] M. A. Albahar, \"Skin lesion classification using convolutional neural network with novel Regularizer,\" in Proc. IEEE Int. Symp. Biomed. Imaging (ISBI), 2019. [16] Yu et al., \"Hybrid dermoscopy image classification framework based on deep convolutional neural network and fisher vector,\" in Proc. IEEE Int. Symp. Biomed. Imaging (ISBI), 2017.
Copyright
Copyright © 2024 Sharadindu Adhikari, Tanmay Kumar Agrawal, Soumyadip Mondal, Ayushmaan Agarwal. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET64032
Publish Date : 2024-08-21
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

