Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Comparative Analysis and Drug Target Identification of Human Kidney Cancer
Authors: Anurag Tripathi, Uma Kumari
DOI Link: https://doi.org/10.22214/ijraset.2023.55777
Certificate: View Certificate
Abstract
Kidney cancer, particularly renal cell carcinoma (RCC), is a significant global health concern characterized by its high morbidity and mortality rates. In this study, we employ a multifaceted approach to gain insights into the molecular mechanisms underlying kidney cancer and to identify potential drug targets for therapeutic intervention. To achieve this, we conducted a comprehensive comparative analysis of gene expression profiles in normal and cancerous kidney tissues, utilizing tools such as BLAST and gene analysis from the COSMIC database. This analysis allowed us to identify key genetic alterations and pathways associated with kidney cancer progression. Furthermore, molecular docking simulations were performed to predict potential drug candidates that could target specific proteins implicated in kidney cancer development. The aim was to prioritize molecules with high binding affinity and therapeutic potential. Additionally, cobalt analysis was utilized to investigate the metalloprotein content within kidney cancer cells, shedding light on potential metalloprotein targets for drug development.The results of this study provide valuable insights into the molecular basis of kidney cancer, offering a foundation for the identification of novel drug targets and the development of targeted therapies for this life-threatening disease. These findings contribute to our understanding of kidney cancer pathogenesis and may ultimately improve patient outcomes and the overall management of this challenging malignancy.
Introduction
I. INTRODUCTION
Renal cell carcinoma, also known as hypernephroma or Grawitz tumor, stands as the predominant form of kidney cancer, constituting over 90% of renal malignancies in the adult population[1].Renal malignancy is a complex group of diseases, comprised of various cancer types categorized by their histological characteristics. These distinct cancers exhibit differences in their presentation, clinical progression, and genetic origins. In 2016, it was projected that around 62,700 new instances of kidney cancer would be identified in the United States, leading to 14,200 fatalities and a consistent rise in occurrence over the past ten years[2]. It represents a diverse collection of cancers originating from the epithelial cells of renal tubules, accounting for approximately 85% of all primary kidney tumors [3].Amongst RCC, the primary subtypes consist of clear cell, papillary and chromophobe RCC. In the realm of kidney tumors, the residual 15% encompasses a diverse range of conditions, including transitional cell carcinoma, nephroblastoma, rare collecting duct tumors, elusive renal sarcomas, and the infrequent occurrence of renal medullary carcinomas [4]. Age, ethnicity, and sex are additional factors that influence the dynamics of this ailment. RCC predominantly affects males beyond the age of 60, with the median age for RCC diagnosis being 65. The zenith of its incidence occurs within the 6th to 8th decades of one's life [5].
The predominant genetic anomaly linked to the pathogenesis of ccRCC is the deletion of the of 3p arm. This genetic aberration is prevalent in nearly 95% of the cases [6]. The genesis of RCC can be attributed to a myriad of influencing factors, which can be classified into two categories: genetic predisposition and acquired risk elements. Genetic factors include: the Von Hippel-Lindau (VHL) gene emerges as a central figure, functioning as a formidable tumor suppressor gene. It is noteworthy that alterations or mutations within the VHL gene are discernible in approximately 50-60% of sporadic ccRCC cases, thus underlining its pivotal role in the etiology of this particular renal cancer subtype [7].It's a recurring theme that modifications in the PBRM-1 gene often find their place in this cancer as well [8]. Several other players take the stage, including SETD2, BAP-1, KDM5C, and MTOR genes [9]. On the other hand, acquired risk factors include smoking, hypertension, obesity, diabetes and chronic analgesic use.Glutaminase is an enzyme responsible for facilitating the transformation of glutamine into glutamate and ammonia. Its pivotal function lies in the glutaminolysis process, wherein glutamine is broken down to supply energy and metabolic intermediates for diverse cellular functions, including the growth and multiplication of cancer cells [10]. Three variants of human glutaminase have been recognized, namely the kidney-type glutaminase (KGA), the splice KGA variant (also known as Glutaminase C or GAC), and the liver-type glutaminase (LGA or GLS2) [11].
The KGA isoform is predominantly found in the kidney, where it plays a role in overseeing glutamine metabolism in this organ. Additionally, its expression has been noted to increase in specific cancer cells, where it plays a part in the heightened glutaminolysis and energy generation necessary to support tumor expansion [12, 13].
The KGA gene exhibits markedly elevated expression levels in clear cell renal cell carcinoma (ccRCC), which is the prevalent form of kidney cancer, in contrast to its expression in healthy kidney tissue. Consequently, the suppression of KGA activity by means of a small molecule inhibitor resulted in a decline in cell viability and an augmentation in cell death among ccRCC cells. The primary focus of this study was to conduct an extensive examination of the KGA protein. In pursuit of this objective, a multifaceted approach was employed, combining various bioinformatics tools to unravel critical insights. The methodology encompassed sequence alignment and bioinformatics analyses, executed through advanced computational techniques such as BLAST. Furthermore, the study delved into the intricate realm of protein-ligand interactions using state-of-the-art Autodock software, which enabled a profound exploration of the molecular dynamics and potential therapeutic targets. This multifaceted approach enhances our understanding of the genetic underpinnings of renal cancer and provides a foundation for the development of targeted therapies in the future.
II. MATERIALS AND METHODS
In the realm of bioinformatics, computer programs serve as fundamental tools that emulate the intricate functions of biological information systems.
These programs undertake complex computations, supplying biologists with invaluable insights into DNA sequences, protein structures, gene regulation mechanisms, and various biological phenomena. The foundation of bioinformatics lies in the utilization of computational software, enabling researchers to delve deep into the biological world. This particular study harnessed the resources of the MMDB database and the National Center for Biotechnology Information (NCBI) to access essential sequences. NCBI stands as a comprehensive repository of biotechnology and biomedicine-related databases, offering an array of bioinformatics tools and services.
Furthermore, to establish sequence similarity, BLASTp was employed. The alignment of multiple sequences was executed using Cobalt, while the COSMIC software was instrumental in deciphering the amino acid composition of protein sequences. Additionally, the study involved the retrieval of ligands from PubChem and target 3D structures from the Protein Data Bank (PDB), with subsequent examination using PYMOL to visualize interactions.
These ligands were then subjected to docking simulations through specialized software, and the resulting outcomes were meticulously observed and analyzed, enhancing our understanding of critical biological interactions.CBDock is a docking web server is an online platform that allows researchers to perform molecular docking simulations. Molecular docking is a computational technique used in drug discovery and structural biology to predict the binding interactions between a small molecule (ligand) and a biological target (typically a protein receptor).
These interactions are crucial for understanding how potential drugs or compounds may bind to a target protein and modulate its activity. PyMOL is a widely used and highly versatile molecular visualization software program primarily designed for the visualization and analysis of three-dimensional (3D) molecular structures, with a particular emphasis on proteins, nucleic acids, and small molecules.
Developed by Warren L. DeLano, PyMOL has become a valuable tool in various scientific disciplines, including structural biology, biochemistry, drug discovery, and molecular modeling.While PyMOL itself is primarily a visualization tool, it can be integrated with other software for molecular docking simulations. Researchers often use PyMOL to visualize and analyze the results of docking studies[14, 15, 16, 17].
III. RESULTS AND DISCUSSION
Following the acquisition of the protein sequence corresponding to the Kidney isoform of Glutaminase from the MMDB database via the National Center for Biotechnology Information (NCBI), the sequence is then converted into the FASTA format. Subsequently, it is employed as the search query in the BLAST tool.
BLAST functions by analyzing the given protein sequence through comparison with a comprehensive database encompassing various protein sequences. The results of the BLASTP search yielded an extensive list of sequences in the descriptions that exhibited similarity to the queried sequence.


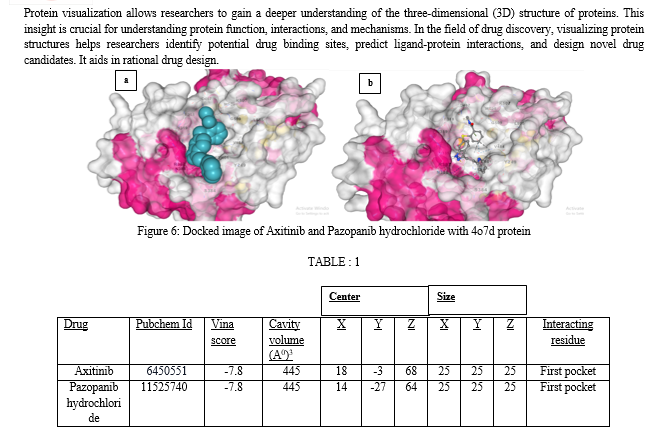
The interacting residues present in the first pocket are TYR249, PHE318, ASN319, ASN335, SER384, ARG387, ASN388, TYR466, ASP467, VAL484, LEU505, LYS507, MET508, GLY509.
Axitinib, marketed under the trade name Inlyta, is a potent and selective small-molecule tyrosine kinase inhibitor (TKI) used in the treatment of advanced renal cell carcinoma (RCC), a type of kidney cancer. Axitinib exerts its therapeutic effects by targeting and inhibiting specific receptors known as VEGFRs, particularly VEGFR-1, VEGFR-2, and VEGFR-3. These receptors play a crucial role in the growth of blood vessels (angiogenesis) that supply tumors with nutrients and oxygen. By inhibiting these receptors, Axitinib interferes with angiogenesis, thereby reducing the blood supply to the tumor and slowing down its growth.Pazopanib hydrochloride, commonly marketed under the brand name Votrient, is a targeted cancer therapy that belongs to the class of drugs known as tyrosine kinase inhibitors (TKIs). It is primarily used in the treatment of advanced renal cell carcinoma (RCC), a type of kidney cancer, and advanced soft tissue sarcoma (STS), a rare form of cancer that affects the soft tissues of the body.
Conclusion
The comprehensive study on kidney cancer, specifically renal cell carcinoma (RCC), has shed light on critical aspects of this formidable disease. Kidney cancer\'s high morbidity and mortality rates underscore the urgent need for deeper insights and therapeutic solutions. Through a multifaceted approach that encompassed comparative gene expression analysis, molecular docking simulations, and metalloprotein investigation, we have made significant strides in understanding the molecular underpinnings of kidney cancer. Our gene expression analysis unveiled pivotal genetic alterations and signaling pathways associated with kidney cancer progression. This knowledge not only enhances our comprehension of the disease\'s mechanisms but also provides potential biomarkers for diagnosis and prognosis.The molecular docking simulations allowed us to identify promising drug candidates with a high binding affinity to specific proteins implicated in kidney cancer development. These candidates hold the promise of becoming the foundation for targeted therapies, offering hope to patients with limited treatment options. Additionally, our cobalt analysis uncovered insights into the role of metalloproteins within kidney cancer cells. This knowledge opens up new avenues for drug development, particularly in targeting metalloprotein-based vulnerabilities within the disease.Collectively, our findings represent a significant step forward in our battle against kidney cancer. They provide the research community with valuable data and avenues for further investigation. Ultimately, the identification of novel drug targets and the development of tailored therapies offer renewed optimism for improving patient outcomes and advancing the overall management of this challenging malignancy. Our dedication to the fight against kidney cancer remains unwavering, driven by the hope of translating these discoveries into tangible clinical benefits for those affected by this devastating disease.
References
[1] Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020 Jan;70(1):7-30. [2] Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. [3] Hsieh JJ, Purdue MP, Signoretti S, et al.: Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. [4] Chow WH, Dong LM, Devesa SS: Epidemiology and risk factors for kidney cancer. Nat Rev Urol. 2010;7(5):245–57. [5] Thompson RH, Ordonez MA, Iasonos A, et al.: Renal cell carcinoma in young and old patients - is there a difference? J Urol. 2008;180(4):1262–6. [6] Beroukhim R, Brunet JP, Di Napoli A, et al.: Patterns of gene expression and copy-number alterations in von-hippellindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69(11):4674–81. [7] Foster K, Prowse A, van den Berg A, et al.: Somatic mutations of the von Hippel-Lindau disease tumour suppressor gene in non-familial clear cell renal carcinoma. Hum Mol Genet. 1994;3(12):2169–73. [8] Thompson M: Polybromo-1: the chromatin targeting subunit of the PBAF complex. Biochimie. 2009;91(3):309–19. [9] Zoncu R, Efeyan A, Sabatini DM: mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. [10] Masisi BK, El Ansari R, Alfarsi L, Rakha EA, Green AR, Craze ML. The role of glutaminase in cancer. Histopathology. 2020 Mar;76(4):498-508. [11] Jin L, Alesi GN, Kang S. Glutaminolysis as a target for cancer therapy. Oncogene. 2016 Jul 14;35(28):3619-25. [12] Aledo, J. C., Gomez-Fabre, P. M., Olalla, L. & Marquez, J. Identification of two human glutaminase loci and tissue-specific expression of the two related genes. Mamm Genome 11, 1107–1110 (2000). [13] Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu Rev Nutr. 1995;15:133-59. [14] Uma Kumari, Nitika kumari,Devendra Kumar, Insilico analysis and computer Aided Drug Designing approach for mutant Cancer gene, 2021/10/21, IJBTR, Volume11, Issue2, Page 21-28 [15] Dr Uma kumari, Tanishkauttam, computational Analysis comparison prediction of Anti-cancer peptide, 2022/5, Journal IJRASET, Volume 10, Issue 6, Pages 3316-3319 [16] Uma Kumari, Shruti Gupta, NGS and Sequence Analysis with Biopython for Prospective Brain Cancer Therapeutic Studies, https://doi.org/10.22214/ijraset.2023.50885 [17] Dr Uma kumari, Devanshi Gupta, In silico RNA aptamer drug design and modelling,2022/4, Journal-JETIR,Volume-9, Issue-4, Pages 718-725
Copyright
Copyright © 2023 Anurag Tripathi, Uma Kumari. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET55777
Publish Date : 2023-09-18
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

