Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
ECG Classification using Deep Learning
Authors: Vishnu Vardhan Atmakuri, Nanda sri Mogili, Naseer Hussain Mohammed, Anusha Neela, Om Vyshanavi Singamsetty, Aqil Hussain Mohammed
DOI Link: https://doi.org/10.22214/ijraset.2024.59181
Certificate: View Certificate
Abstract
The importance of ECG categorization has grown significantly, as demonstrated by the many contemporary medical applications that address this issue. A plethora of machine learning (ML) tools are already available for the analysis and classification of ECG data. The primary drawbacks of these machine learning outcomes are the utilization of heuristic hand-crafted or manufactured features with shallow feature learning architectures, or the acquisition of numerical data using image processing. The use of CNN, which recognizes and captures specific patterns in images, is one suggested solution for this problem utilizing deep learning architecture. Recurrent neural networks (RNN) and their variant LSTM are advantageous in the ECG classification task due to their implicit capacity to effectively capture temporal dependencies and noise while providing interpretability of hidden states. In contrast to human results, these Deep Learning Techniques yield results about the employment of Recurrent Neural Networks with LSTM layers and Convolutional Neural Networks without any feature-engineered procedure and competing classification accuracy.
Introduction
I. INTRODUCTION
The use of deep learning algorithms has been a game-changer in the field of cardiovascular imaging, allowing for the extraction of intricate features and patterns from ECG data, resulting in highly accurate classification of various cardiac conditions. This article aims to explore the effectiveness of convolutional neural networks and recurrent neural networks with long short-term memory layers in ECG classification, highlighting their ability to capture temporal dependencies and noise while offering interpretability of hidden states. Additionally, we will discuss the advantages of deep learning approaches over traditional machine learning solutions, which often rely on heuristic hand-crafted features or shallow learning architectures. Through an extensive analysis of more than 300 research articles in the medical field, it has been determined that deep learning techniques, particularly convolutional neural networks and recurrent neural networks with LSTM layers, have shown great promise in the classification of ECG data. These deep learning algorithms have the advantage of not requiring hand-crafted or engineered features, as they can automatically learn and extract relevant features from the raw ECG data (Romiti et al., 2020). Furthermore, these deep learning techniques have demonstrated superior performance compared to human results, showcasing their ability to accurately classify various cardiac conditions without the need for manual intervention.
II. LITERATURE REVIEW
In order to automatically classify ECGs of various rhythms, including AF, Zhaohan Xiong [1] built RhythmNet, a 21-layer 1D convolutional recurrent neural network. The network was trained using 8,528 single lead ECG recordings from the 2017 PhysioNet/Computing in Cardiology (CinC) Challenge. Their RhythmNet architecture consisted of three recurrent layers to process ECGs of different lengths and identify arrhythmia events in lengthy records, after which 16 convolutions to extract features directly from raw ECG waveforms. To efficiently learn the intricate fluctuations of the signal within short time-frames, like P-waves and QRS complexes, large 15 × 1 convolutional filters were employed.
Roger G. Mark [2], A E Johnson, Qiao Li, Ikaro Silva, Li-wei H. Lehman, Benjamin Moody, and Gari D. Clifford, among others. PhysioNet/Computing in Cardiology Challenge 2017: AF Classification using a Short Single Lead ECG Recording.
Previous research on the classification of AF is typically limited in its applicability because: 1) only the classification of AF and normal rhythms was done, The vast taxonomy rhythms makes it particularly difficult to properly identify AF from a single short lead of the ECG. Specifically, irregular RR intervals that may resemble AF are present in a large number of non-AF rhythms.
Chao Chen [3] Proposed employing a 1-layer LSTM model in conjunction with an 8-layer CNN with eight shortcut connections to identify AF in single lead, brief ECG data.
By adding shortcut connections, can speed up the processing and data transfer of conventional convolutional neural networks. In addition, it has a fully connected layer and a 1-layer LSTM that can handle long-term reliance between data in addition to effectively extracting features. Based on the test set of the Computing in Cardiology Challenge 2017 dataset, the three deep learning models were assessed with an F1 score. Three sets of comparative trials revealed that 8CSL outperformed RNN and MCNN on all indexes.
[4] Another research used preprocessed time-series data and 1D convolutional neural networks with FCN layers to offer some ECG classification results. On validation data, the best accuracy result was around 86%. [5] Moreover, the rule-based methodology and deep learning network approach can be used to automatically classify ECG data. The rule-based approach and the deep learning methods like GoogleNet yielded validation scores of 0.325 and 0.426, respectively, based on the time-frequency and morphological ECG features with labels.
A diagnostic method for Myocardial Infarction, a cardiovascular condition, was proposed by Acharya et al [6]. Because of this, they constructed a novel method for detecting normal and abnormal ECGs with and without noise, and they were able to attain accuracy rates of 95.22% without noise removal and 93.53% with noise. Throughout the leads, a voting system was used to identify the arrhythmia in several datasets. The ECG signals are transformed into scaleograms before being trained using the transfer learning SqueezeNet model for picture categorization. As a result, the entire test score is 0.205 and the achieved validation score is 0.214, respectively [7]-[9]. With the use of feature reduction and a rule-based fuzzy classifier, cardiovascular disease was predicted with an accuracy of 76.51% in an experiment conducted on UCI datasets [10]. In addition, the bioimaging-based machine learning algorithm appears to be beneficial in the identification of breast cancer, which is thought to have a high global death rate [11].
Artificial intelligence (AI) is a commonly utilized technique in the treatment of colorectal cancer sickness. It works with class-selective image processing to detect cancer in the colon and lungs by applying transfer learning and scaled dilation in CNN [12] [13]. It is believed that the existing approaches do not function sufficiently well. The 1-D Convolutional Neural Network, or 1-D CNN, is quick, simple, and effective. For cardiac arrhythmic diseases, the 1-D CNN's accuracy was 91.33%, and its classification time was 0.015 seconds [14].
III. METHODOLOGIES
A. CNN Model
The entire document should We used Deep Learning techniques such as CNN, RNN with LSTM layers and proposed two models where first model is actually CNN and secondly we combined CNN with RNN. This model architecture starts with a Convolutional Neural Network (CNN) component, consisting of three Conv2D layers. The first Conv2D layer has 32 filters with a filter size of 3x3 and utilizes the Rectified Linear Unit (ReLU) activation function. It takes input images with a shape of 150x150 pixels and 3 color channels, representing RGB. Following the Conv2D layer, a MaxPooling2D layer with a pool size of 2x2 is applied to downsample the feature maps. The figure 1 shows the CNN model architecture.
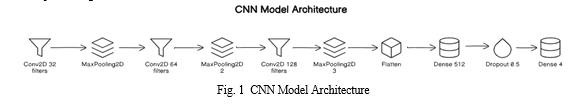
Subsequently, another Conv2D layer with 64 filters and a filter size of 3x3 is employed, followed by another MaxPooling2D layer with the same pool size. After that, the model includes a third Conv2D layer with 128 filters and a filter size of 3x3, followed by a MaxPooling2D layer with a pool size of 2x2. These convolutional layers aim to extract hierarchical features from the input ECG images, capturing both low-level and high-level patterns. Following the convolutional layers, the model utilizes a Flatten layer to reshape the output from the convolutional layers into a one-dimensional vector. This flattening operation is necessary to transition from the spatial understanding of the features in the convolutional layers to the fully connected layers. After flattening, the model incorporates two Dense (fully connected) layers for classification. The first Dense layer consists of 512 neurons and applies the ReLU activation function to learn high-level features from the flattened input. A Dropout layer with a dropout rate of 0.5 is added after the first Dense layer to prevent overfitting by randomly dropping a fraction of input units during training. Finally, the model concludes with the output layer, a Dense layer with four neurons representing the four classes in the classification task. The softmax activation function is applied to this layer, producing a probability distribution over the classes. This architecture aims to effectively learn and extract features from ECG images for accurate classification into the respective classes.
B. CNN-RNN Model
The figure 2 shows the architecture of proposed CRNN model. Our proposed model combines convolutional layers (Conv2D and MaxPooling2D) for feature extraction with recurrent layers (LSTM) for sequential temporal data processing. While the LSTM layers evaluate the sequential information produced by the convolutional layers, convolutional layers capture spatial features from the input images. The model architecture integrates Convolutional Neural Network (CNN) and Recurrent Neural Network (RNN) components to process Electrocardiogram (ECG) images efficiently for classification. It begins with three Conv2D layers followed by MaxPooling2D layers, progressively increasing the number of filters from 32 to 128. Each Conv2D layer uses a 3x3 filter size and Rectified Linear Unit (ReLU) activation function, processing input images of 150x150 pixels and 3 color channels (RGB). Subsequently, MaxPooling2D layers down sample the feature maps to capture the most salient features. After the convolutional layers, a Time Distributed layer is applied to flatten the feature maps, preparing them for input into the Recurrent Neural Network (RNN) layers. Two Long Short-Term Memory (LSTM) layers follow, with the first LSTM layer containing 128 units and configured to return sequences.

A dropout layer with a 0.3 dropout rate is inserted after the first LSTM layer to mitigate overfitting. Following the LSTM layers, a second LSTM layer with 128 units further captures temporal patterns in the ECG signals. Two Dense (fully connected) layers are then added for classification, with the first layer comprising 512 neurons and employing the ReLU activation function. A dropout layer with a 0.4 dropout rate is placed after this layer to enhance generalization. Finally, the output layer consists of four neurons representing the classification classes, employing the softmax activation function to produce class probabilities.
IV. IMPLEMENTATION
The Dataset consists of 928 ECG images of Normal beat(284), Abnormal beat(233), Myocardial Infarction(239) and History of MI(172), is divided into train, validation and test sets, then based on ratios ( 70% for training, 15% for validation, and 15% for testing), the images are allocated to their respective sets. The number of images assigned to each set is proportional to the specified ratios. We preprocessed the images before using the dataset of raw 12 Lead ECG report images to train the model. Before even starting the preprocessing steps first we check the dimensions of the image, if they match with the dataset images dimensions it goes to preprocessing or else if it does not match it will display a message saying that Uploaded image does not have suitable dimensions for preprocessing.
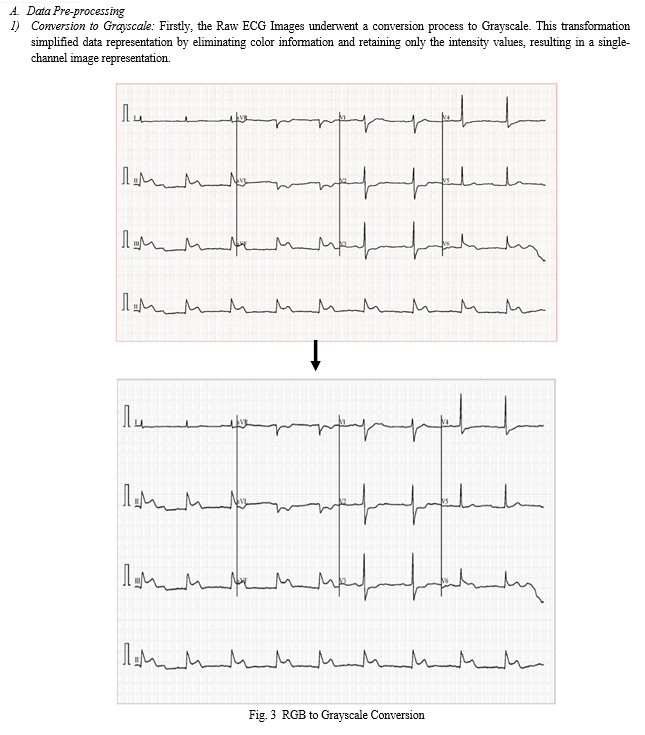

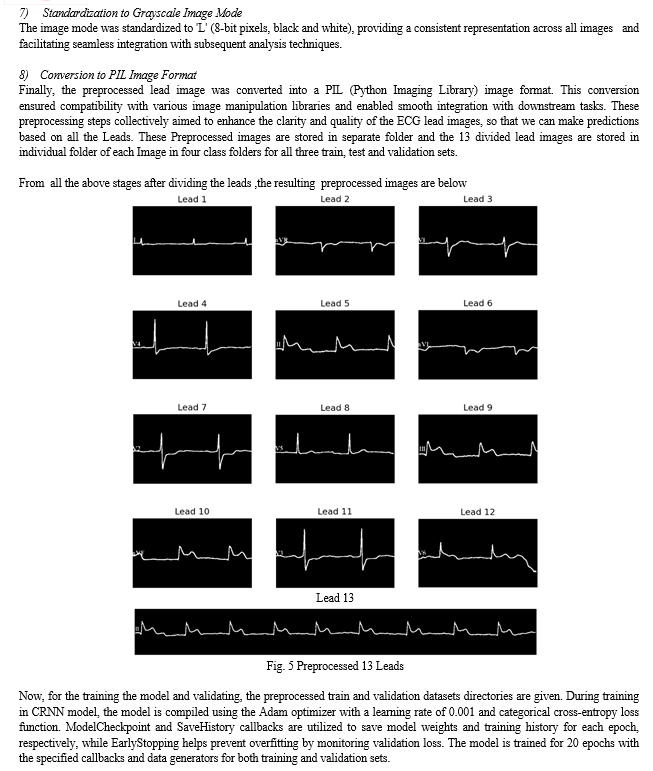
B. Classification Process
Upon launching the application, users are prompted to upload an ECG image from their local system. Once an image is uploaded, it is displayed in the user interface, providing users with a visual representation of the uploaded image. The application then proceeds to preprocess the uploaded image, a crucial step in preparing the image for classification. During preprocessing, the uploaded image undergoes several transformations, including conversion to grayscale and division into individual leads. These preprocessing steps are essential for extracting relevant features from the ECG image and facilitating accurate classification. The grayscale version of the image and its divided leads are displayed in expandable sections within the user interface, allowing users to inspect the preprocessing results visually. After preprocessing, the application employs a pre-trained deep learning models (CNN, CRNN) to classify the ECG image. The preprocessed leads are resized to a standard size and fed into the model for prediction. The model predicts the class probabilities for each lead, and these predictions are displayed in the interface. The predicted class probabilities for each lead are displayed, providing insights into the model's confidence in its predictions for individual leads. Additionally, the overall predicted class for the ECG image is determined based on the aggregated predictions from all leads. This information is presented to the user, enabling them to understand the classification outcome and make informed decisions based on the model's predictions. All things considered, the application provides an interactive and intuitive platform for the classification of ECG images, enabling users to efficiently examine and understand their ECG data.
V. RESULTS
The model performed well on unseen data, and the classification results of unseen test data also shown good results. Both the CNN and CRNN models demonstrated strong performance in training and validation stages, showcasing their ability to effectively classify ECG images. Their precision, recall, and F1-score metrics indicate their reliability in accurately identifying patterns in heart activity. These results highlight the models' potential in contributing significantly to medical diagnosis and advancing the field of ECG image classification.
The CNN model demonstrated exceptional performance during training, achieving an accuracy of 97.82%. Even on unseen validation data, it maintained accuracy of 93.40%. Moreover, its precision stands at 93.45%, with a recall of 93.38% and an overall F1-score of 93.31%. Meanwhile, the proposed CRNN model exhibited a training accuracy of 98.82% and a validation accuracy of 93.72%. Notably, it achieved a precision of 93.74%, matching its recall rate of 93.72%, resulting in a robust F1-score of 93.70%. These results underscore the effectiveness of both models in accurately classifying ECG images, promising significant contributions to the field.


VI. LIMITATIONS
- We can consider some strategies to improve validation accuracy compared to training accuracy.
- Although the model is trained with 12,064 lead images, the dataset consists of in total 928 raw ECG images only.
- The model works well with the dataset only, which we have taken and even if we upload an image which is different from ECG image it classifies as abnormal heartbeat which has to be in same dimensions as the image as in the dataset, although it has a dimension tolerance of 15 pixels.
VII. FUTURE WORK
Additionally, we can fine-tune model complexity and hyperparameters through techniques like cross-validation. These adjustments aim to enhance the model's ability to generalize to unseen data, ensuring more reliable performance in real-world scenarios. To enhance the dataset's coverage beyond the existing four classes used, we can explore collaborations with healthcare institutions for diverse ECG recordings or employ data synthesis techniques to simulate additional classes, enabling the model to better generalize across a broader spectrum of cardiac abnormalities.
VIII. ACKNOWLEDGEMENT
- We would like to express our heartfelt appreciation to the authors whose work has been cited in this article. Their invaluable contributions have significantly enriched our understanding of the subject matter and have been instrumental in shaping the direction of our research.
- We would want to convey our gratitude for all of the help and advice we received in finishing this paper. We would like to thank our project coordinator Dr. N. Sri Hari and project guide Mr. A. Vishnu Vardhan for their help and Essential guidance during the whole work.
Conclusion
Since Artificial intelligence has made a significant impact on human life, it is essential to many real-world industries, including finance, healthcare, and medicine. In the current generation, AI subdomains like machine learning and deep learning are regarded as leading technologies. The field of medicine makes extensive use of deep learning techniques like image segmentation and classification. This work has put into practice a paradigm that addresses a broad area of healthcare, namely the classification of ECG reports. For automatic and precise findings, the proposed paradigm can be used in medical diagnosis equipment such as embedded systems. Through this project, we have demonstrated the effectiveness of deep learning models, such as convolutional neural networks (CNNs) and recurrent neural networks (RNNs), in accurately classifying ECG signals, which are crucial for diagnosing various cardiac conditions. These automation-related actions will accelerate the healthcare sector\'s rapid growth and prepare the way for a better future.
References
[1] Convolutional Recurrent Neural Network-Based ECG Signal Classification for the Identification of Cardiac Arrhythmias. Vadim V. Fedorov, Elizabeth Cheng, Zhaohan Xiong, Martyn P. Nash, Martin K. Stiles, and Jichao Zhao. [2] PhysioNet/Computing in Cardiology Challenge 2017: AF Classification using a Short Single Lead ECG Recording, Roger G. Mark, A E Johnson, Qiao Li, Ikaro Silva, Li-wei H. Lehman, Benjamin Moody, and Gari D. Clifford, among others. [3] CNN-LSTM and Shortcut Connection-Based Automatic Atrial Fibrillation Detection Yongjie Ping, Chao Chen, Lu Wu, Yinglong Wang, and Minglei Shu , Shandong University of Science and Technology, Qingdao 266590, China; pyjboxmail@163.com (Y.P.); lwu@qlu.edu.cn (L.W.). College of Computer Science and Engineering. [4] Deep Learning for ECG Classification B Pyakillya et al 2017 J. Phys.: Conf. Ser. 913 012004. [5] “Rule-based approach and deep learning networks for automatic ECG categorization”, G. Bortolan, I. Christov, and I. Simova, Proceedings of the 2020 Computing in Cardiology, pp. 1-4, IEEE, Rimini, Italy, 2020, September. [6] \"Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals,\" Information Sciences, vol. 415–416, pp. 190–198, 2017, by U. R. Acharya, H. Fujita, S. L. Oh, Y. Hagiwara, J. H. Tan, and M. Adam. [7] Automated classification of electrocardiograms using wavelet analysis and deep learning, A. Demonbreun and G. M. Mirsky, Proceedings of the 2020 Computing in Cardiology, pp. 1-4, IEEE, Rimini, Italy, September 2020. [8] Reduced-lead ECG classification using wavelet analysis and deep learning, A. K. Cornely, A. Carrillo, and G. M. Mirsky, Proceedings from the 2021 Computing in Cardiology (CinC), pp. 1-4, IEEE, Brno, Czech Republic, September 2021. [9] \"SE-ECGNet: multi-scale SE-Net for multi-lead ECG data,\" J. Chen, T. Chen, B. Xiao, et al., Proceedings of the 2020 Computing in Cardiology, pp.1-4, IEEE, Rimini, Italy, September 2020. [10] In the Proceedings of the International Conference on Machine Learning, pp. 3597–3606, PMLR, Chennai, India, November 2020, T. Golany, K. Radinsky, and D. Freedman, \"Simgans: simulator-based generative adversarial networks for ECG synthesisto improve deep ECG classification.\" [11] \"Bio-imagingbased machine learning algorithm for breast cancer detection,\" Diagnostics, vol. 12, no. 5, p. 1134, 2022, S. Safdar, M. Rizwan, T. R. Gadekallu, et al. [12] In 2020, IEEE Access published a paper titled \"Colorectal disease classification using efficiently scaled dilation in convolutional neural network,\" authored by S. Poudel, Y. J. Kim, D. M. Vo, and S. W. Lee. [13] S. Mehmood, T. M. Ghazal, M. A. Khan, et al., \"Using transfer learning with class selective image processing for malignancy detection in lung and colon histopathology images,\" IEEE Access, vol. 10, Article ID 25657, 2022. [14] O Y?ld?r?m, P. P?awiak, R. S. A. Tan, and U. R. Acharya “Arrhythmia detection using deep convolutional neural network with long duration ECG signals,” Computers in Biology and Medicine, vol. 102, pp. 411–420, 2018.
Copyright
Copyright © 2024 Vishnu Vardhan Atmakuri, Nanda sri Mogili, Naseer Hussain Mohammed, Anusha Neela, Om Vyshanavi Singamsetty, Aqil Hussain Mohammed. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET59181
Publish Date : 2024-03-20
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

