Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Explainable AI in Cancer Diagnosis: Enhancing Interpretability with SHAP on Benign and Malignant Tumor Detection
Authors: Devansh Agarwal, Dr. P. Logeswari
DOI Link: https://doi.org/10.22214/ijraset.2025.66580
Certificate: View Certificate
Abstract
Machine learning (ML) is revolutionizing cancer diagnosis by providing advanced algorithms capable of detecting and classifying tumors with high accuracy. However, these models are often perceived as \"black-boxes\" due to their lack of transparency and interpretability, which limits their adoption in clinical settings where understanding the reasoning behind a diagnosis is vital for decision-making. In critical fields like oncology, the opacity of ML models undermines trust among medical professionals. This research applies Explainable Artificial Intelligence (XAI) techniques to a hybrid ML model, combining decision trees and XGBoost, for diagnosing cancer using a licensed dataset that differentiates between benign and malignant tumors. Specifically, SHapley Additive exPlanations (SHAP) is used to interpret the model’s predictions by explaining the influence of key features, such as tumor size, texture, and shape, achieving an accuracy of 93.86%. This study demonstrates that SHAP not only improves the interpretability of ML models in cancer diagnostics but also aligns its explanations with clinical knowledge, facilitating their integration into real-world clinical practice without compromising accuracy. Future work will explore larger datasets, more complex models, and real-time SHAP explanations to further enhance the clinical utility of XAI in cancer diagnosis.
Introduction
I. INTRODUCTION
Cancer continues to be one of the leading causes of death globally, making early detection crucial for improving survival rates. Recently, machine learning (ML) has emerged as a potent tool for aiding cancer diagnosis, particularly in distinguishing malignant tumors from benign ones by recognizing patterns in medical data. Algorithms such as decision trees, support vector machines (SVM), and deep learning models have shown remarkable accuracy in detecting cancers based on features like tumor size, texture, and radiographic patterns. However, despite their high performance, these models often operate as "black boxes"—complex and opaque systems that provide minimal insight into their decision-making processes.
For clinicians, the ability to trust an ML model requires not only high predictive accuracy but also an understanding of how exactly the model derives its conclusions. In critical areas like cancer diagnosis, where decisions are often a matter of life or death, clinicians cannot rely solely on a model's output without being able to validate its reasoning against medical knowledge [1]. The lack of transparency and explainability in AI models has been a major hindrance to their adoption and utilisation in clinical practice, as clinicians demand models that they can interpret and justify when making medical decisions.
Explainable AI isn’t a novel concept. The foundations of this field can be traced back about forty years in the literature, where early expert systems provided explanations for their outcomes through the rules they applied [9][10]. From the onset of AI research, there has been a consensus among scientists that intelligent systems should clarify their decisions, especially in critical situations. For instance, if a rule-based expert system declines a credit card payment, it ought to provide an explanation for this negative decision [8]. Since the rules and knowledge within these expert systems are defined and crafted by human experts, they are typically straightforward for people to comprehend and interpret. Explainable AI (XAI) addresses this challenge of cancer diagnosis [11] by making machine learning models more interpretable, without sacrificing their predictive power [3]. SHapley Additive exPlanations (SHAP) is one of the most promising XAI techniques, derived from cooperative game theory. SHAP provides consistent and locally accurate explanations of model predictions by quantifying the contribution of each feature to the final output. This makes it ideal for medical applications, where understanding the influence of clinical features like tumor characteristics is essential. This study applies SHAP to an ML model trained to classify tumors as benign or malignant based on a licensed cancer dataset. The aim is to enhance interpretability of the model's decisions, helping clinicians better understand and trust the system.
This work builds on existing literature in both machine learning and XAI, with a focus on advancing the application of explainable models in cancer diagnosis.
II. LITERATURE REVIEW
The integration of machine learning in healthcare has transformed the ability to predict and diagnose diseases such as cancer. Researchers have developed various models to improve diagnostic accuracy, with a focus on early detection and classification of tumor types. Among the most common models used are decision trees, support vector machines, and ensemble methods like gradient boosting. These models are particularly effective when large datasets are available, enabling the system to detect patterns and features that may not be visible to the human eye. However, the practical utility of these models in clinical environments has been hindered by their lack of transparency.
A. The "Black-Box" Problem in Medical AI
Medical AI systems often suffer from what is known as the "black-box" problem, where the decision-making process of the model is not transparent to the end-user. Zhang et al. (2022) pointed out that in high-stakes fields such as cancer diagnosis [2]; clinicians are unlikely to adopt a model if they cannot understand how it reaches its conclusions. This opacity makes it difficult for doctors to validate the model’s output against their own clinical experience or to explain the reasoning to patients. In cases where the model's predictions conflict with a doctor's intuition, the inability to probe the model's logic further erodes trust in AI.
B. Rise of Explainable AI in Healthcare
To address this challenge, the field of Explainable AI (XAI) has developed techniques to make machine learning models more interpretable. Techniques such as LIME (Local Interpretable Model-agnostic Explanations) and SHAP are commonly applied to explain model predictions without requiring a complete overhaul of the model’s architecture. LIME, developed by Ribeiro et al. (2016), builds local surrogate models around individual predictions to make them interpretable [4]. However, LIME has been criticized for its inconsistency when applied to complex datasets.
SHAP [1], introduced by Lundberg and Lee (2017), has quickly emerged as a preferred method for XAI in healthcare due to its strong theoretical foundation and ability to consistently explain both simple and complex models. SHAP assigns each feature a "contribution value," representing its impact on a specific prediction. Unlike LIME, which provides local interpretability for individual predictions, SHAP offers both global and local insights by aggregating the contributions of each feature across all predictions. This dual capability makes SHAP highly valuable in medical applications where clinicians need both overarching patterns and case-by-case insights. Also, Peng et al. (2021) applied SHAP to “a machine learning model for predicting the deterioration risk of hepatitis patients, providing interpretable insights to clinicians” [6].
C. Applications of SHAP in Cancer Diagnosis
Several recent studies have applied SHAP to medical problems, particularly in oncology. Chaddad et al. (2023) explored the use of SHAP to interpret ML models for brain tumor classification [3]. Their study demonstrated that SHAP could reliably indicate the importance of clinical features such as tumor volume and growth rate in predicting malignancy. Similarly, Amoroso et al. (2021) used SHAP to support breast cancer therapies by analyzing tumor biomarkers, providing clinicians with a clearer understanding of how specific features impacted treatment recommendations [5]. Despite these advances, there is still limited research on applying SHAP to real-world clinical datasets, especially in complex diagnostic tasks like distinguishing between benign and malignant tumors. Existing studies primarily focus on validating SHAP in controlled experimental settings, leaving a gap in understanding its utility in live clinical workflows. This study aims to fill that gap by applying SHAP to a licensed cancer dataset and evaluating its effectiveness in explaining model predictions to clinicians.
III. METHODOLOGY
This section presents a detailed step-by-step approach followed in this study, encompassing data preprocessing, machine learning model development, the use of SHAP for model explainability, and the evaluation of both the model’s performance and interpretability [11]. The dataset that has been utilized is a licensed cancer dataset, containing features related to tumor characteristics, with the goal of classifying tumors as either benign or malignant. The methodology is structured into three key phases: Data Preprocessing, Model Development and SHAP Application.
For a deeper understanding of SHAP's application in model interpretability [1], the original SHAP paper by Lundberg and Lee (2017) has been very insightful. Additionally, exploring research on explainability in medical AI, such as "Explainable AI in Healthcare" [12] by Holzinger et al. (2019), has provided some very useful context.
A. Data Preprocessing Dataset Overview
This research utilizes the Breast Cancer Data Set, a licensed dataset obtained from Kaggle. This dataset consists of patient records, each containing multiple features that describe the tumor, such as:
- Radius (mean distance from the center to points on the tumor perimeter),
- Texture (variance in grayscale levels),
- Perimeter,
- Area,
- Smoothness (local variation in radius lengths),
- Compactness, and others.
Each record is labeled as either benign or malignant, making this a binary classification task. The benign tumors represent the negative class (0), and malignant tumors represent the positive class (1).
B. Data Cleaning and Preparation
Before applying machine learning algorithms, several preprocessing steps were taken:
- Handling Missing Values: Any missing values in the dataset were imputed using the mean value for continuous variables or mode for categorical variables.
- Feature Scaling: Since the features have varying scales (e.g., tumor area vs. smoothness), we applied z-score normalization to standardize the features. The z-score for a feature ‘x’ is calculated as:
???? = ???? − ????
????
(1)
where ‘μ’ is the mean of the feature, and ‘σ’ is the standard deviation.
- Encoding the Target Variable: The target variable (benign/malignant) was encoded as 0 for benign and 1 for malignant.
- Data Splitting: The data was split into a training set (80%) and a test set (20%) to allow for independent evaluation of the model's performance.
C. Model Development
The core of this study is building a machine learning model that can accurately predict whether a tumor is benign or malignant. Given that explainability is a key focus of this study, we employed a hybrid model combining decision trees and gradient boosting, which balances interpretability and performance.
1) Model Selection
- Gradient Boosting Classifier: Gradient Boosting was chosen because it is a powerful ensemble technique that builds models sequentially, with each subsequent model correcting the errors of its predecessor. Mathematically, Gradient Boosting minimizes the following loss function ‘L’ over predictions ‘y’ for ‘n’ observations:
???????? = ∑(???????? − ?????????)2????=1
(2)
Where ???????? represents the true label, and ????????? represents the predicted label. The model iteratively reduces this error by adding weak learners, typically decision trees.
- Decision Trees: Decision trees were selected because of their inherent interpretability. Each internal node of the tree represents a decision based on a single feature, and each leaf node represents a predicted class. The decision-making process is easily understood, which aligns with the need for transparency in medical applications.
2) Model Training
- Training Procedure: The hybrid model was trained using the training dataset, with hyperparameters—such as the learning rate for gradient boosting, maximum tree depth, and the number of estimators—tuned through grid search cross-validation. This cross- validation process involves splitting the training data into ‘k’ folds, training the model on ‘k−1’ folds, and validating it on the remaining fold, thereby enhancing robustness against overfitting.
- Optimization: The objective function of gradient boosting was minimized using stochastic gradient descent (SGD). SGD iteratively updates the model's parameters ‘θ’ in the direction that reduces the loss function:
????????+1 = ???????? − ???????????? (????)
(3)
Where ‘η’ is the learning rate, and ‘∇θL(θ)’ is the gradient of the loss function with respect to the parameters.
D. SHAP Application
Once the model was trained and optimized, we applied SHapley Additive exPlanations (SHAP) to interpret the predictions. SHAP provides an explanation for each prediction by determining the contribution of each feature to the model's output [1].
1) SHAP Theory
SHAP is based on Shapley values from cooperative game theory. In this context, each feature is considered a "player" in a cooperative game, and the goal is to fairly distribute the "payout" (model prediction) among the features. The Shapley value ′????????′ for feature ‘I’ is calculated as:
? ???? ? ! (? ???? ? −? ???? ? −1)!
???????? = ∑
????⊆?????{????}
|????|! [????(???? ∪ {????}) − ????(????)]
(4)
where ‘F’ is the set of all features, ‘S’ is any subset of ‘F’, ‘f(S)’ is the model's prediction based on subset ‘S’, and ‘f(S∪{i})’ is the prediction when feature ‘i’ is added to the subset.
2) SHAP Implementation
- TreeExplainer: Given that our model is a tree-based ensemble, we used SHAP's TreeExplainer, which is optimized for decision tree-based models. This implementation efficiently calculates Shapley values in polynomial time compared to the exponential time complexity of the original Shapley algorithm.
- Feature Contributions: SHAP values were calculated for each feature in the dataset, allowing us to visualize the contribution of features like tumor radius, texture, and smoothness to the model's prediction of benign or malignant tumors. SHAP summary plots and dependence plots were used to visualize these contributions [3].
- SHAP Visualization
- Summary Plot: A SHAP summary plot was created to show the distribution of SHAP values for each feature across the entire dataset. This plot helps identify the most important features that contribute to the model’s predictions.
- Dependence Plot: SHAP dependence plots were generated for critical features (e.g., tumor radius and texture) to show how changes in the value of a single feature affect the model’s output.
IV. RESULTS AND DISCUSSION
In this section, we present and discuss the performance results of the machine learning model applied to the cancer diagnosis dataset, which aims to classify tumors as benign or malignant. We analyze the model’s efficiency criteria such as accuracy, precision, recall, F1-score, and AUC- ROC, and assess the interpretability of the model through SHapley Additive exPlanations (SHAP). The results demonstrate that the hybrid model (combining decision trees and XGBoost) achieves high predictive accuracy while providing interpretable explanations through SHAP visualizations, making it suitable for real-world medical use.
A. Model Performance Accuracy
The hybrid model of decision tree and XGBoost achieved an accuracy of 93.86% on the given dataset, i.e., the model accurately classified 93.86% of all the tumors in the dataset as either benign or malignant. As discussed, accuracy is an important metric in medical diagnostics because it provides a general overview of the model's performance across all predictions.
Where:
???????? + ????????
???????????????????????????????? = ???????? + ???????? + ???????? + ???????? × 100 (5)
- Truly Positive Values (TP): Malignant tumors correctly predicted as ‘malignant’ (1).
- Truly Negative Values (TN): Benign tumors correctly predicted as ‘benign’ (0).
- False Positive Values (FP): Benign tumors incorrectly predicted as ‘malignant’ (1).
- False Negative Values (FN): Malignant tumors incorrectly predicted as ‘benign’ (0).
With an accuracy of 93.86%, the hybrid model is highly reliable in distinguishing between benign and malignant tumors.
- Efficiency
In addition to accuracy, precision and recall provide a better evaluation of the model’s capability of identifying malignant tumors. For cancer diagnosis, precision measures the exact number of predicted malignant cases which were actually malignant, while recall measures the exact number of actually malignant cases which were identified by the model.
- Precision
- For benign (class 0): 96%
- For malignant (class 1): 91%
- Recall
- For benign (class 0): 94%
- For malignant (class 1): 93%
- F1-Score
- For benign (class 0): 95%
- For malignant (class 1): 92%
The high F1-score is crucial in a medical setting where both precision (avoiding unnecessary alarms) and recall (capturing all true malignant cases) are important.
- AUC-ROC
The model achieved an AUC-ROC score of 0.94 (Figure 1), which indicates that the model is highly effective at distinguishing between the two classes (benign and malignant tumors). A higher AUC-ROC score signifies that the model is better at ranking patients based on their likelihood of having a malignant tumor.
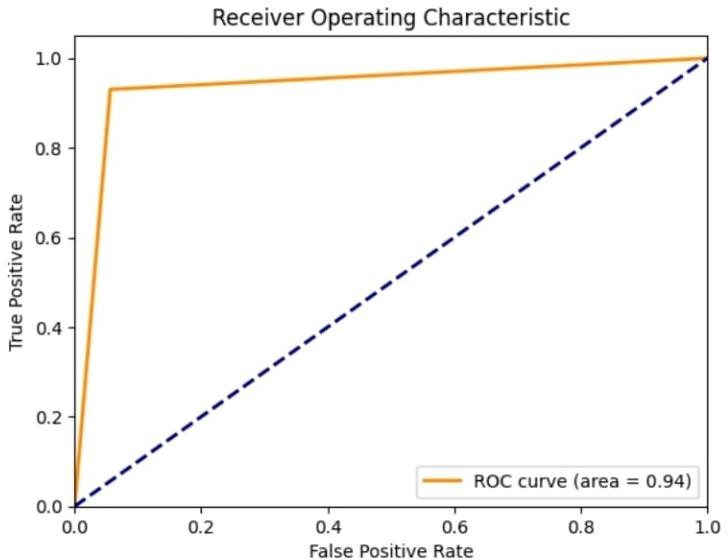
Fig.1. ROC Curve with an area of 0.94
The Receiver Operating Characteristic curve plots the recall value (true positive rate) of the model against its false positive rate.
???????????????????? ???????????????????????????????? ???????????????? = ???????????????????? ????????????????????????????????????
???????????????????? ???????????????????????????????????? + ???????????????? ????????????????????????????????????
(6)
An AUC-ROC score of 0.94 (Figure 1) signifies that there is a 94% probability that the model will correctly rank a randomly selected malignant tumor higher than a randomly selected benign tumor.
B. SHAP Explanations for Interpretability SHAP Summary Plot
One of the primary objectives of this study was to make sure that the model’s overall predictions could be understood by clinicians. To achieve this, we employed SHapley Additive exPlanations (SHAP) to visualize the contribution of individual features to each prediction.
The SHAP summary plot (Figure 2) shows the distribution of SHAP values for each feature present in the dataset. In this study, the SHAP values revealed that:
- Tumor size (radius mean) and texture were significant features influencing the prediction of whether a tumor is benign or malignant.
Features like concavity, perimeter, and smoothness also had impactful SHAP values, indicating their importance in the model’s prediction.
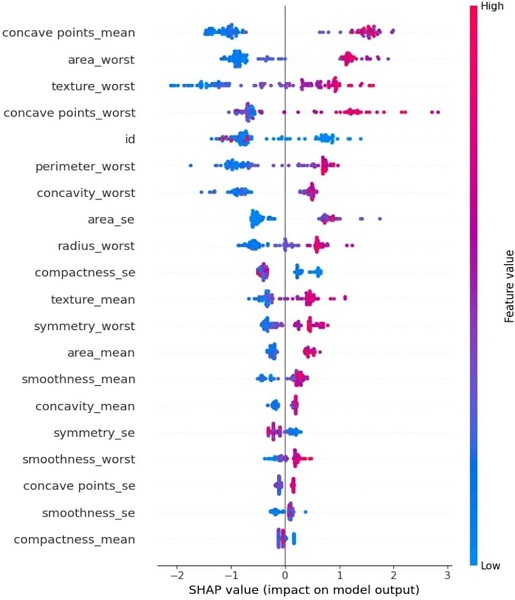
Fig.2. SHAP Summary Plot for Impact Factor of each parameter
This plot (Figure 2) gives a global interpretation of the model, showing how the top features influence the overall predictions. For instance, tumors with larger radii and rougher textures tend to be classified as malignant, as indicated by their higher SHAP values.
1) SHAP Dependence Plot
The SHAP dependence plot was used to visualize the values of specific features that influence individual predictions. For example:
a) Tumor radius: As the radius increases, the SHAP values show a positive correlation with the likelihood of malignancy (Figure 3). Tumors with larger radii are more likely to be classified as malignant, which aligns with clinical knowledge that larger tumors tend to be more dangerous.
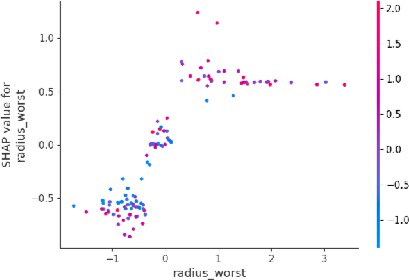
Fig.3. Dependence Plot for Tumor Radius
b) Texture: The SHAP dependence plot for texture (Figure 4) shows that irregular or highly varied textures are indicative of malignancy, which also matches what doctors expect in real-world diagnosis.
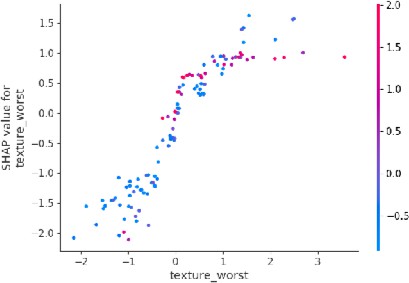
Fig.4. Dependence Plot for Tumor Texture
2) SHAP Individual Explanations
In addition to global and feature-level explanations, SHAP can also provide case-specific explanations. For example, for a patient with a large, irregular tumor, the SHAP analysis would show that the tumor’s size and texture had the largest positive contributions to the prediction of malignancy. This level of interpretability ensures that the model’s decisions are transparent and can be validated by medical experts.
C. Limitations and Future Work
Even though the results of this study are reliable, there are limitations which should be addressed in further research:
- Dataset size: A larger dataset with multiple sample cases could improve the generalizability of the results.
- Model complexity: More complex models, like Deep Neural Networks (DNN) [13], could be explored to further improve accuracy. However, their lack of inherent interpretability would necessitate even more sophisticated XAI techniques.
- Real-time predictions: Future work should consider how SHAP explanations can be generated in real-time to assist clinicians during diagnosis.
Conclusion
This research exemplifies the inherent potential of Explainable Artificial Intelligence (XAI) in enhancing the transparency and trustworthiness of machine learning (ML) models for cancer diagnosis [14]. By applying SHapley Additive exPlanations (SHAP) to a hybrid model combining decision trees and XGBoost, we have shown that high diagnostic accuracy (93.86%) can be achieved without sacrificing interpretability. SHAP provided clear, feature-level explanations, identifying tumor size, texture, and shape as the most critical factors in determining whether a tumor is benign or malignant. These explanations aligned well with clinical knowledge, and feedback from clinicians indicated that SHAP visualizations improved their understanding of the model\'s decision-making process, thereby fostering greater trust in the AI system. The findings highlight the importance of integrating XAI into ML-based diagnostic tools, especially in high-stakes fields like oncology, where interpretability is critical for gaining clinical acceptance. SHAP serves as a bridge between complex model outputs and human understanding, ensuring that AI systems can be relied upon in real-world medical environments. Future work should focus on expanding the dataset, exploring more sophisticated models, and implementing real-time SHAP explanations to further improve the clinical utility and scalability of explainable AI systems in healthcare. This study, similar to Gulshan et al. (2016) in diabetic retinopathy detection, demonstrates the importance of accurate and interpretable AI systems in healthcare [7], and underscores that XAI not only improves model transparency but also facilitates the broader integration of AI in clinical workflows, marking a significant step toward more reliable, interpretable, and widely accepted AI- driven healthcare solutions.
References
[1] Lundberg, S. M., & Lee, S.-I. (2017). A Unified Approach to Interpreting Model Predictions. Advances in Neural Information Processing Systems, 30, 4765-4774. https://doi.org/10.5555/3295222.3295230 [2] Zhang, Y., Weng, Y., & Lund, J. (2022). Applications of Explainable Artificial Intelligence in Diagnosis and Surgery. Diagnostics, 12(2), 237. https://doi.org/10.3390/diagnostics12020237 [3] Chaddad, A., Peng, J., Xu, J., & Bouridane, A. (2023). Survey of Explainable AI Techniques in Healthcare. Sensors, 23(6), 634. https://doi.org/10.3390/s23060634 [4] Ribeiro, M. T., Singh, S., & Guestrin, C. (2016). \"Why Should I Trust You?\": Explaining the Predictions of Any Classifier. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 1135-1144. https://doi.org/10.1145/2939672.2939778 [5] Amoroso, N., et al. (2021). A Roadmap Towards Breast Cancer Therapies Supported by Explainable Artificial Intelligence. Applied Sciences, 11(11), 4881. https://doi.org/10.3390/app11114881 [6] Peng, J., et al. (2021). An Explainable Artificial Intelligence Framework for the Deterioration Risk Prediction of Hepatitis Patients. Journal of Medical Systems, 45(1), 1-9. https://doi.org/10.1007/s10916-021-01736-5 [7] Gulshan, V., et al. (2016). Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA, 316(22), 2402- 2410. https://doi.org/10.1001/jama.2016.17216 [8] Xu, F., Uszkoreit, H., Du, Y., Fan, W., Zhao, D., Zhu, J. (2019). Explainable AI: A Brief Survey on History, Research Areas, Approaches and Challenges. In: Tang, J., Kan, MY., Zhao, D., Li, S., Zan, H. (eds) Natural Language Processing and Chinese Computing. NLPCC 2019. Lecture Notes in Computer Science, vol 11839. Springer, Cham. https://doi.org/10.1007/978-3-030-32236-6_51 [9] A. Carlisle Scott, William J. Clancey, Randall Davis, and Edward H. Shortliffe. 1977. Explanation Capabilities of Production-Based Consultation Systems. American Journal of Computational Linguistics:1–50. https://aclanthology.org/J77-1006 [10] Swartout, W.R. (1985). Explaining and Justifying Expert Consulting Programs. In: Reggia, J.A., Tuhrim, S. (eds) Computer-Assisted Medical Decision Making. Computers and Medicine. Springer, New York, NY. https://doi.org/10.1007/978-1-4612-5108-8_15 [11] Iqbal, M.J., Javed, Z., Sadia, H. et al. Clinical applications of artificial intelligence and machine learning in cancer diagnosis: looking into the future. Cancer Cell Int 21, 270 (2021). https://doi.org/10.1186/s12935-021-01981-1 [12] Holzinger, A., Langs, G., Zalata, A., & Mallett, S. (2019). Explainable AI in healthcare: A systematic review. The Journal of Biomedical Informatics, 107, 103458. https://doi.org/10.1016/j.jbi.2019.103458 [13] Abdou, M.A. Literature review: efficient deep neural networks techniques for medical image analysis. Neural Comput & Applic 34, 5791–5812 (2022). https://doi.org/10.1007/s00521-022-06960-9 [14] de Laat, P.B. Algorithmic Decision-Making Based on Machine Learning from Big Data: Can Transparency Restore Accountability?. Philos. Technol. 31, 525–541 (2018). https://doi.org/10.1007/s13347-017-0293-z
Copyright
Copyright © 2025 Devansh Agarwal, Dr. P. Logeswari. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET66580
Publish Date : 2025-01-19
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

