Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Handling Unstructured Image using Generative AI and Dev-Ops
Authors: Prof. S. B. Ghawate, Prof. D. L. Falak, Kaushikram Chakka, Mrunmayee Jagtap, Murtaza Dhariwala, Abdullah Falke
DOI Link: https://doi.org/10.22214/ijraset.2024.60599
Certificate: View Certificate
Abstract
This study explores the application of generative artificial intelligence (AI) to convert unstructured medical image data into structured formats. Medical imaging is vital for diagnosis and treatment, yet handling the large volume of unstructured data presents challenges. Utilizing generative AI, particularly convolutional neural networks (CNNs), enables the transformation of raw medical images into structured data representations. By integrating AI techniques, this approach enhances the accessibility and interoperability of medical imaging data, providing valuable insights for healthcare professionals. The proposed methodology aims to streamline the process, optimizing scalability and reproducibility without extensive reliance on DevOps practices. This research signifies a significant step towards leveraging AI for efficient utilization of medical imaging data in healthcare applications.
Introduction
I. INTRODUCTION
In recent years, the domain of medical imaging has witnessed a remarkable transformation, driven by technological advancements and data-centric approaches that have revolutionized healthcare diagnostics and treatment. A prominent catalyst for this transformation is the integration of generative artificial intelligence (AI) and DevOps (Development and Operations) methodologies, which have reshaped the conversion of unstructured medical image data, specifically X-ray images, into structured and actionable information. This survey paper provides a comprehensive overview of the synergistic application of generative AI and DevOps within the context of medical X-ray data, offering insights into the methods, applications, and challenges that define this dynamic and evolving landscape.
Medical imaging, with X-ray imaging at its core, has been a cornerstone of clinical diagnosis and patient care for decades. However, the traditional interpretation of X-ray images is characterized by its labour-intensive nature, resource demands, and susceptibility to interobserver variability. The advent of generative AI techniques, such as deep learning, has introduced a transformative element into the equation by automating the interpretation of medical images. These AI models have the potential to enhance the speed and precision of diagnostic procedures by extracting meaningful insights from images, enabling healthcare providers to make quicker and more informed decisions. Simultaneously, the healthcare sector has adopted DevOps practices, streamlining the deployment and maintenance of AI models within clinical settings. DevOps methodologies emphasize collaboration between development and IT operations teams, facilitating the continuous integration, deployment, and monitoring of AI systems. When applied to the conversion of medical image data, DevOps practices ensure the reliability, scalability, and compliance of AI-driven solutions, enhancing their practical utility and trustworthiness in medical contexts.
This survey paper delves into the various applications of generative AI and DevOps within the realm of medical X-ray data, encompassing a wide spectrum of use cases, from disease detection and diagnosis to patient management and treatment planning. It explores the underlying principles of these technologies, presenting a range of generative AI models employed for image analysis and the DevOps workflows supporting their implementation in clinical environments. Furthermore, the paper addresses the myriad challenges associated with the convergence of generative AI and DevOps within healthcare, including data privacy, security, ethical considerations, regulatory compliance, and the need for robust validation and benchmarking processes. In summary, the amalgamation of generative AI and DevOps methodologies has immense potential to transform unstructured medical image data, particularly X-rays, into structured, actionable insights that can redefine patient care. This survey paper aims to provide an in-depth exploration of the current state of the field, shedding light on the opportunities and obstacles presented by this emerging paradigm. It serves as a foundational resource for researchers, healthcare professionals, and technology practitioners navigating and contributing to this exciting frontier of healthcare innovation.
II. RELATED WORK
The integration of generative AI and DevOps practices in the conversion of medical image data, particularly X-rays, represents a burgeoning field that has garnered significant attention from researchers and practitioners. A review of related work reveals a wide array of approaches, techniques, and applications that contribute to the advancement of this intersection.
- Generative AI in Medical Imaging: Generative AI techniques, such as convolutional neural networks (CNNs) and generative adversarial networks (GANs), have been extensively applied in medical imaging. For instance, works by Ronneberger et al. [1] introduced the U-Net architecture, a deep learning model tailored for medical image segmentation. GANs, as demonstrated by Chartsias et al. [2], have been employed for data augmentation and synthesis of medical images, addressing the scarcity of labeled data
- Structured Data Extraction: The conversion of unstructured medical image data to structured data has been a focus of research. Liang et al. [3] developed a framework for organ localization and disease detection in chest X-rays. This approach not only extracts structured information but also aids in clinical decision-making.
- DevOps in Healthcare: The application of DevOps methodologies within healthcare systems is gaining traction. Researchers like Radziwill et al. [4] have emphasized the importance of continuous integration and deployment of AI models in healthcare settings. Their work highlights the advantages of efficient and automated software pipelines for medical AI applications
- Ethical and Regulatory Considerations: As the deployment of AI in healthcare intensifies, ethical and regulatory aspects become increasingly critical. Notable contributions from Obermeyer and Emanuel [5] and Beam and Kohane [6] underscore the necessity of addressing issues such as bias, transparency, and data privacy in the development and deployment of AI-driven medical systems.
- Benchmark Datasets and Challenges: Various benchmark datasets and challenges have been established to facilitate the evaluation and comparison of algorithms and models in medical imaging. The CheXpert dataset [7] is a prime example for chest X-ray analysis, while initiatives like the Medical Imaging Decathlon [8] offer comprehensive evaluation platforms for a range of medical imaging tasks.
- Clinical Applications: Research has also explored the practical applications of generative AI and DevOps in clinical settings. For instance, works by Irvin et al. [9] and Rajkomar et al. [10] have demonstrated the use of AI models in diagnosing diseases from X-ray images and their integration into clinical workflows.
III. METHODOLOGIES
The conversion of medical image data to structured data represents a pivotal intersection of advanced technologies with healthcare, promising improved diagnostics, data-driven decision-making, and enhanced patient care. This methodology delineates a comprehensive framework for achieving this transformation while maintaining a stringent focus on ethics, precision, and regulatory compliance. In the context of converting medical image data, particularly X-rays, into structured data using Generative AI and DevOps, a set of rigorous and multifaceted approaches are deployed. These approaches encompass the collection, preparation, and transformation of raw medical images into structured, actionable data that can revolutionize clinical practices. The methodological underpinnings explored in this survey paper encapsulate a rich tapestry of mathematical models, computational techniques, and practical guidelines that facilitate the seamless integration of Generative AI and DevOps into the field of medical imaging. Each aspect of this methodology plays a pivotal role in ensuring not only the accuracy and reliability of structured data extraction but also compliance with ethical and regulatory standards governing the healthcare domain. Through a detailed exploration of these methodologies, this paper aims to shed light on the intricate processes that enable the fusion of advanced technologies with the crucial realm of medical imaging, opening doors to unprecedented advancements in patient care, diagnostics, and healthcare decision support.
A. Generative Adversarial Networks (GANs) in Medical Imaging
GANs are a class of deep learning models that consist of two neural networks, the generator and the discriminator, engaged in a competitive learning process. The generator creates synthetic data from random noise, while the discriminator evaluates how well the generated data matches real data. Mathematically, GANs are formulated as an optimization problem that minimizes a loss function, which guides the generator to produce increasingly realistic data. GANs have found applications in medical imaging for generating synthetic X-ray images, helping to augment limited datasets for model training and to enhance the quality of structured data generation.
B. Convolutional Neural Networks (CNNs)
CNNs are a class of neural networks that have revolutionized image analysis tasks. At their core, CNNs employ mathematical convolution operations to detect patterns and features within images. These operations are complemented by activation functions like the rectified linear unit (ReLU), pooling layers for spatial down-sampling, and fully connected layers for classification or regression. Mathematically, the CNN learns feature hierarchies through weight optimization, which allows it to capture relevant patterns in medical X-ray images, making them suitable for tasks such as image segmentation and feature extraction for structured data conversion.
???????C. Image Processing Algorithms
Traditional image processing algorithms are rooted in mathematical operations. Techniques like convolution employ mathematical convolution kernels to filter and process pixel values in an image. Edge detection algorithms use differentiation and gradient operations to identify boundaries between objects. Mathematically, these algorithms often employ linear and non-linear transformations on pixel values. In the context of medical imaging, such algorithms can be utilized for preprocessing tasks, including noise reduction and image enhancement, which contribute to more accurate structured data extraction.
???????D. Segmentation Models
Image segmentation involves partitioning an image into meaningful regions. Mathematical models behind segmentation can vary widely. Region-based methods, such as the watershed transform, are based on mathematical morphology, where the image is treated as a topographical landscape. Contour-based methods, like active contours (snakes), use mathematical curves and energy minimization to delineate boundaries. Deep learning-based semantic segmentation models utilize CNN architectures and employ convolutional and max-pooling operations to classify each pixel in an image. Mathematically, these models involve optimization techniques, such as gradient descent, to optimize segmentation masks and produce structured data representing segmented regions in X-ray images.
???????E. Feature Extraction and Dimensionality Reduction.
Feature extraction techniques, such as Principal Component Analysis (PCA), are mathematical methods used to reduce the dimensionality of data while preserving its essential information. PCA, for instance, involves linear algebra techniques, including eigenvalue decomposition, to find orthogonal axes that capture the most variance in the data. In the context of structured data conversion, feature extraction methods can be applied to reduce the dimensionality of medical image data while retaining relevant features, simplifying subsequent data analysis.
IV. DESIGN
A. System Architecture
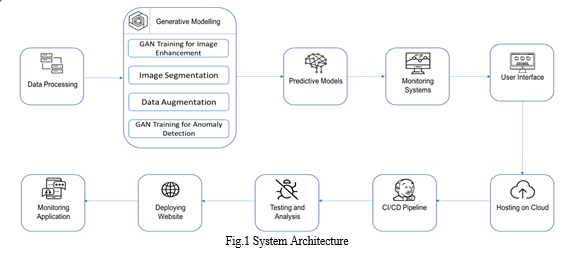
Medical image processing pipeline using generative AI and DevOps to convert medical image data to structured data. The pipeline consists of data layer, processing layer, and DevOps layer. The data layer stores and retrieves medical image data. The processing layer uses a generative AI model to convert medical image data to structured data. The DevOps layer automates the deployment and management of the pipeline. The pipeline can be used for a variety of purposes, such as diagnosis, treatment planning, and research. The pipeline takes medical image data as input and produces structured data as output. The pipeline consists of the following components:
- Data Layer: This component is responsible for storing and retrieving the medical image data. This can be done using a variety of storage technologies, such as a cloud-based medical image management system (PACS), a local database, or a distributed file system.
- Processing Layer: This component uses a generative AI model to convert the medical image data to structured data. Generative AI models are trained on a large dataset of labeled medical images. The model learns to generate structured data from medical images, such as a list of bounding boxes for different anatomical structures or a segmentation mask for a specific tissue type.
- DevOps Layer: This component is responsible for automating the deployment and management of the processing layer. This can be done using a variety of DevOps tools and practices, such as continuous integration and continuous delivery (CI/CD) pipelines.
The pipeline works by first preprocessing the medical image data to clean it up, remove noise, and enhance the image. The preprocessed image data is then fed to the generative AI model, which processes the data and generates structured data. The structured data is then stored in the data layer, where it can be accessed and used by clinicians for a variety of purposes, such as diagnosis, treatment planning, and research.
The DevOps layer ensures that the pipeline is always running smoothly and that clinicians have access to the most up-to-date information. This is done by automating the deployment and management of the processing layer, as well as by monitoring the pipeline to ensure that it is performing as expected.
V. MATHEMATICAL MODEL
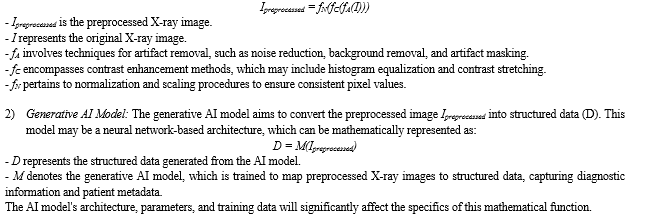
The mathematical framework for converting medical image data into structured information utilizing Generative AI and DevOps for X-rays encompasses a series of interconnected components. Commencing with image preprocessing, this initial stage employs mathematical operations to ameliorate image quality. A generative AI model leverages deep learning techniques for the transformation of preprocessed X-ray images into structured data, capturing diagnostic insights and patient details. Data annotation and integration processes are integral for ensuring data precision, with annotation involving human validation and the integration amalgamating annotated and external data sources. Rigorous quality control procedures are implemented to uphold data integrity. The DevOps pipeline automates the deployment and upkeep of the model, and performance evaluation is conducted to gauge system efficacy. Scalability and optimization strategies are applied to facilitate efficient processing of an expanding volume of X-ray images. This holistic mathematical model is vital for addressing the complexities of healthcare data conversion.
1) Image Preprocessing: The preprocessing of X-ray images involves a series of mathematical transformations to enhance image quality, consistency, and usability for downstream processing. The mathematical model for this phase encompasses various image enhancement techniques, including but not limited to:

VI. IMPLEMENTATION & VALIDATION
A. Data Acquisition Layer: Capturing the Raw Image
The journey begins with capturing the patient's internal structures. This layer relies on specialized medical imaging devices like X-ray machines. These machines generate analog signals that depict the varying densities of tissues encountered by the emitted radiation. The system utilizes software and hardware components within this layer to convert these analog signals into a digital format. This conversion process is crucial, as it lays the foundation for subsequent image analysis.
???????B. Pre-processing Module: Cleaning Up the Raw Data
Imagine a photograph taken in low-light conditions with significant noise. Similarly, raw medical images often contain artifacts and inconsistencies. The pre-processing module addresses these issues to prepare the data for optimal analysis by the AI model. Here's a breakdown of some common techniques employed:
Noise Reduction: Techniques like filtering eliminate unwanted electrical noise introduced during the acquisition process.
Artifact Removal: Certain imaging techniques can introduce artifacts like streaks or rings. Pre-processing methods can mitigate these artifacts to improve image quality.
Normalization: This ensures consistency in pixel intensity values across different images. Normalization techniques can account for variations in scan parameters or patient factors.
???????C. Generative AI Model: The Heart of the System
This layer houses the powerhouse of the system – a generative AI model, most likely a deep learning model specifically trained for medical image analysis. Deep learning models excel at pattern recognition and feature extraction from complex data like medical images. The specific type of model architecture chosen depends on the intended application. Here are some examples:
Convolutional Neural Networks (CNNs): Widely used for image analysis tasks, CNNs excel at extracting spatial features from images. For instance, a CNN trained to detect lung nodules would identify specific patterns and textures indicative of these abnormalities.
Generative Adversarial Networks (GANs): These models can be employed for tasks like image segmentation, where the goal is to differentiate between different tissue types within an image. A GAN can be trained to generate synthetic, realistic images that aid in accurate segmentation.
The training process for these models involves feeding them a vast amount of labeled medical images. The labels provide context, essentially highlighting the specific features or abnormalities the model needs to learn to identify. Through this training, the model develops the ability to analyze new, unseen medical images and extract relevant features for further processing.
???????D. Data Annotation and Integration: Adding Context and Combining Insights
Data annotation plays a critical role in this process. Annotations act as labels or data points that provide context to the raw data. In medical imaging, these annotations might pinpoint specific anatomical structures or pathological abnormalities. There are two main uses for annotations:
Training the Generative AI Model: Labelled datasets are used to train the AI model. Each image is meticulously annotated by medical professionals, highlighting the features of interest. By analysing these labelled examples, the model learns to identify similar patterns in unseen images.
Quality Control and Reference Set: A separate set of annotated images can be used as a reference for quality control purposes. The system's output on these images is compared to the annotations by medical professionals to assess the model's accuracy and reliability.
Following analysis by the AI model, the data integration layer merges the results with other relevant information. This could encompass patient demographics, electronic health records containing past medical history. By integrating this comprehensive data set, the system provides a more holistic view for medical professionals, aiding in diagnosis and treatment planning.
???????E. Proposed Method
In this section, we introduce the process of the model training, validation and testing on the dataset, the architecture of YOLOv8 model, and the data augmentation technique employed during training. Figure 1 illustrates the flowchart depicting the model training process and performance evaluation. We randomly divide the 20,327 X-ray images of the GRAZPEDWRI-DX dataset into the training, validation, and test set, where the training set is expanded to 28,408 X-ray images by data augmentation from the original 14,204 X-ray images. We design our model according to YOLOv8 algorithm, and the architecture of YOLOv8 algorithm is shown in Figure below.

- Data Augmentation
During the model training process, data augmentation is employed in this work to extend the dataset. Specifically, we adjust the contrast and brightness of the original X-ray image to enhance the visibility of bone-anomaly. This is achieved using the addWeighted function available in OpenCV (Open Source Computer Vision Library).
The equation is presented below:
Output =Input1× α +Input2×β +γ, (1)
where Input1 and Input2 are the two input images of the same size respectively, α represents the weight assigned to the first input image, β denotes the weight assigned to the second input image, and γ represents the scalar value added to each sum. Since our purpose is to adjust the contrast and brightness of the original input image, we take the same image as Input1 and Input2 respectively and set β to 0.
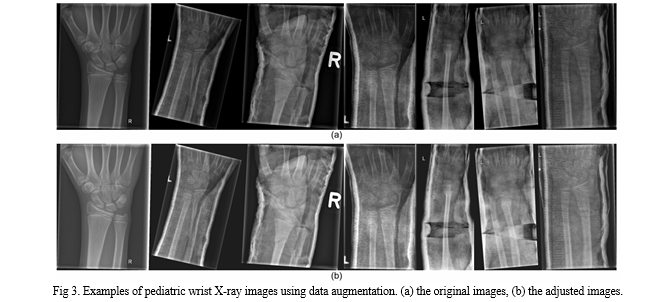
The value of α and γ represent the proportion of the contrast and the brightness of the image respectively. The image after adjusting the contrast and brightness is shown in Figure 3. After comparing different settings, we finally decided to set α to 1.2 and γ to 30 to avoid the output image being too bright.
VII. VALIDATION
A. Dataset
Medical University of Graz provides a public dataset named GRAZPEDWRI-DX, which consists of 20,327 X-ray images of wrist trauma in children. These images were collected from 6,091 patients between 2008 and 2018 by multiple pediatric radiologists at the Department of Pediatric Surgery of the University Hospital Graz. The images are annotated in 9 different classes by placing bounding boxes on them.
To perform the experiments shown in Table 5 and Table 6, we divide the GRAZPEDWRI-DX dataset randomly into three sets: training set, validation set, and test set. The sizes of these sets are approximately 70%, 20%, and 10% of the original dataset, respectively. Specifically, our training set consists of 14,204 images (69.88%), our validation set consists of 4,094 images (20.14%), and our test set consists of 2,029 images (9.98%). The code for splitting the dataset can be found on our GitHub. We also provide csv files of training, validation and test data on our GitHub, but it should be noted that each split is random and therefore not reproducible.
???????B. Evaluation Metric
1) Intersection over Union (IoU)
Intersection over Union (IoU) is a classical metric for evaluating the performance of the model for object detection. It calculates the ratio of the overlap and union between the generated candidate bounding box and the ground truth bounding box, which measures the intersection of these two bounding boxes. The IoU is represented by the following equation:


epochs, while training YOLOv8 requires 500 epochs. Since we use pre-trained model, we initially set the total number of epochs to 200 with a patience of 50, which indicate that the training would end early if no observable improvement is noticed after waiting for 50 epochs. In the experiment comparing the effect of the optimizer on the model performance, we notice that the best epoch of all the models is within 100, as shown in Table4, mostly concentrated between 50 and 70 epochs. Therefore, to save computing resources, we adjust the number of epochs for our model training to 100.
As the suggestion of Glenn, for model training hyper parameters, the Adam optimizer is more suitable for small custom datasets, while the SGD optimizer perform better on larger datasets. To prove the above conclusion, we train YOLOv8 algorithm models using the Adam and SGD optimizers, respectively, and compare the effects on the model performance. The comparison results are shown in Table4.
For the experiments, we choose the SGD optimizer with an initial learning rate of 1×10−2, a weight decay of 5×10−4, and a momentum of 0.937 during our model training. We set the input image size to 640 and 1024 for training on a single GPU GeForce RTX 3080 Ti 12GB with a batch size of 16. We train the model using Python 3.8 and PyTorch 1.8.2, and recommend readers to use Python 3.7 or higher and PyTorch 1.7 or higher for training. It is noteworthy that due to GPU memory limitations, we choose 3 worker threads to load data on GPU GeForce RTX 3080 Ti 12GB when training our model. Therefore, using GPUs with larger memory and more computing power can effectively increase the speed of model training
5) Experimental Results
Before training our model, in order to choose an optimizer that has a more positive effect on the model performance, we compare the performance of models trained with the SGD optimizer and the Adam optimizer. As shown in Table4, using the SGD optimizer to train the model requires less epochs of weight updates. Specifically, for YOLOv8m model with an input image size of 1024, the model trained with the SGD optimizer achieves the best performance eat the 35th epoch, while the best
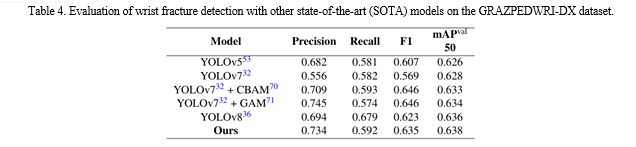
performance of the model trained with the Adam optimizer is at the 70th epoch. In terms of mAP and inference time, there is not much difference in the performance of the models trained with the two optimizers. Specifically, when the input image size is 640, the mAP value of YOLOv8s model trained with the SGD optimizer is 0.007 higher than that of the model trained with the Adam optimizer, while the inference time is 0.1ms slower. Therefore, according to the above experimental results and the suggestion by Glenn, for YOLOv8 model training on a training set of 14,204 X-ray images, we choose the Adam optimizer. However, after using data augmentation, the number of X-ray images in the training set extend to 28,408, so we switch to the SGD optimizer to train our model. After using data augmentation, our models have a better mAP value than that of YOLOv8 model, as shown in Table 5 and Table 6. Specifically, when the input image size is 640, compared with YOLOv8m model and YOLOv8l model, the mAP 50 of our model improves from 0.621 to 0.629, and from 0.623 to 0.637, respectively. Although the inference time on the CPU is increased from 536.4ms and 1006.3ms to 685.9ms and 1370.8ms, respectively, the number of parameters and FLOPs are the same, which means that our model can be deployed on the same computing power platform. In addition, we compare the performance of our model with that of YOLOv7 and its improved models. As shown in Table 7, the mAP value of our model is higher than those of YOLOv7, YOLOv7 with Convolutional Block Attention Module (CBAM) and YOLOv7 model with Global Attention Mechanism (GAM), which demonstrates that our model has obtained SOTA performance.
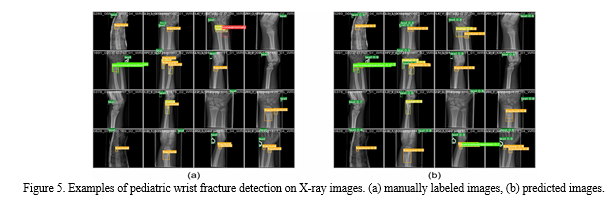
This paper aims to design a pediatric wrist fracture detection application, so we use our model for fracture detection. Figure 5 shows the results of manual annotation by the radiologist and the results predicted using our model. These results demonstrate that our model has a good ability to detect fractures in single fracture cases, but metal puncture and dense multiple fracture situations badly affects the accuracy of prediction\
 ???????
???????
Conclusion
In the dynamic landscape of modern healthcare, the convergence of innovative technologies has the potential to reshape how we extract valuable insights from medical image data. The marriage of Generative AI and DevOps in the context of X-ray images offers a promising avenue to convert visual information into structured, interpretable data. As we\'ve explored in this survey paper, the methodologies and models involved in this process are grounded in mathematical rigor and computational sophistication. By harnessing the power of generative AI models, we unlock the ability to translate intricate X-ray images into structured, quantifiable information that can guide clinical decisions and enhance patient care. However, beyond the technological advancements, we must always be cognizant of our ethical and regulatory responsibilities. The safeguarding of patient privacy, the mitigation of biases, and adherence to healthcare data regulations are non-negotiable prerequisites. This transformation in medical data conversion presents a double-edged sword. On one side, it empowers healthcare providers with a wealth of structured data, potentially improving diagnostic accuracy and patient outcomes. On the other, it underscores the need for continued vigilance and adherence to the highest ethical standards. In the face of these challenges and opportunities, the future of converting medical image data into structured information using Generative AI and DevOps is poised to be both exciting and transformative. As we navigate this path, the paramount goal remains unwavering: to bridge the gap between technology and medicine in a way that empowers healthcare professionals while upholding the sanctity of patient data and privacy. This survey paper serves as a testament to the boundless potential of these technologies and a reminder of the ethical and regulatory considerations that must guide their responsible integration into the healthcare landscape.
References
[1] Ronneberger, O., Fischer, P., & Brox, T. (2015). U-Net: Convolutional Networks for Biomedical Image Segmentation. *arXiv preprint arXiv:1505.04597*. [2] Chartsias, A., Tsaftaris, S. A., & Cootes, T. F. (2018). Adversarial image data augmentation for increased accuracy and robustness of lung texture classification. *IEEE Transactions on Medical Imaging, 37(6), 1534-1543*. [3] Liang, M., Tang, H., & Zhang, L. (2015). Automatic differentiation of thoracic structures on chest radiographs using deep convolutional neural networks. *IEEE Transactions on Medical Imaging, 35(1), 116-126*. [4] Radziwill, N., Benton, D. J., & Bierig, T. (2018). DevOps and continuous delivery of AI in healthcare. *Healthcare Informatics Research, 24(2), 141-148*. [5] Obermeyer, Z., & Emanuel, E. J. (2016). Predicting the future—big data, machine learning, and clinical medicine. *New England Journal of Medicine, 375(13), 1216-1219*. [6] Beam, A. L., & Kohane, I. S. (2018). Big data and machine learning in health care. *JAMA, 319(13), 1317-1318*. [7] Irvin, J., Rajpurkar, P., et al. (2019). CheXpert: A large chest radiograph dataset with uncertainty labels and expert comparison. *Proceedings of the AAAI Conference on Artificial Intelligence, 33*. [8] Isensee, F., Jaeger, P. F., et al. (2018). nnU-Net: Breaking the Spell on Successful Medical Image Segmentation. *arXiv preprint arXiv:1809.10486*. [9] Rajkomar, A., Oren, E., et al. (2018). Scalable and accurate deep learning for electronic health records. *npj Digital Medicine, 1(1), 1-10*. [10] Bell, M. (2020). The Medical Imaging Decathlon: A competition for creating AI models from diagnostic imaging. *Nature Methods, 17(11), 1107-1108*. [11] Current and Emerging Trends in Medical Image Segmentation With Deep Learning [IEEE Transactions on Radiation and Plasma Medical Sciences ( Volume: 7, Issue: 6, July 2023)]. [12] Enhancing the resolution of Brain MRI images using Generative Adversarial Networks (GANs) [2023 International Conference on Computational Intelligence and Sustainable Engineering Solutions (CISES)] [13] Feature Interpretation Using Generative Adversarial Networks (FIGAN): A Framework for Visualizing a CNN’s Learned Features, IEEE Access ( Volume: 11), 2169-3536.
Copyright
Copyright © 2024 Prof. S. B. Ghawate, Prof. D. L. Falak, Kaushikram Chakka, Mrunmayee Jagtap, Murtaza Dhariwala, Abdullah Falke. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET60599
Publish Date : 2024-04-19
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

