Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Heart Failure Prediction Using Machine Learning
Authors: Jeevan Babu Maddala, Bhargav Reddy Modugulla, Sahithi Amulya Pulusu, Sanjay Mannepalli, Praveen prakash Pamidimalla, Rukhiya Khanam
DOI Link: https://doi.org/10.22214/ijraset.2024.59236
Certificate: View Certificate
Abstract
Cardiovascular Disease (CVD) currently stands as the leading cause of death worldwide. Clinical data analytics encounter a significant challenge in accurately predicting cardiac disease. The healthcare industry generates vast volumes of raw data, necessitating its transformation into meaningful insights through machine learning techniques. The objective is to leverage machine learning models to improve the predictability of survival among cardiac patients. This study employs machine learning classifiers: Random Forest, Gradient Boosting classifier, Extra Tree Classifier, XG-Boost, Ada Boost and Hybrid models. The Synthetic Minority Oversampling Technique (SMOTE) addresses the challenge posed by imbalanced datasets. Experimental results indicate that employing the SMOTE technique enhances the accuracy of the chosen classifier\'s predictions. Among these classifiers, Hybrid Model stands out with the highest accuracy of 89.82% when applied to predicting the survival of cardiac illness after implementing SMOTE
Introduction
I. INTRODUCTION
Heart diseases, as per the world health organization (WHO), stand as a predominant cause of mortality worldwide. Identifying cardiovascular disease(CVD) presents challenges due to various contributing factors such as high blood pressure, cholesterol levels, diabetes, abnormal pulse rates, among others. Additionally, symptoms of CVD can vary between genders, where males may predominantly experience chest pain while females may exhibit additional symptoms like nausea, extreme fatigue, and shortness of breath alongside chest discomfort. Despite ongoing research efforts exploring diverse techniques for predicting heart diseases, early-stage disease prediction remains inefficient due to factors like complexity, execution time, and the accuracy of approaches. Effective treatment and diagnosis hold the potential to save numerous lives. Various factors, including blood pressure, cholesterol, and creatinine levels, influence heart health, making diagnosis challenging in the contemporary era, electronic health records (EHRs) play a crucial role in both clinical and research contexts. Minor errors in physical examinations can have signific- ant consequences, especially in cases of heart disease. Machine learning-based expert systems have proven effective in diagnosing CVD, subsequently reducing the mortality rate.
This research primarily focuses on a cardiac dataset comprising 12 input features and 1 output feature, consisting of 300 rows. Upon analysis, no null values were found. In case of null values, they can be replaced with the mean value of the respective feature. Preprocessing of data involved applying SMOTE to balance the dataset based on the output feature, which pertains to mortality events. Following SMOTE, the dataset was augmented to 406 rows with a balanced output feature.
Subsequently, important features were extracted using Random Forest and ETC algorithms, revealing six crucial features: age, creatinine phosphokinase, ejection fraction, serum creatinine, serum sodium, and time. Examination using box plots identified outliers in these features.
Two options were considered for outlier handling: trimming or capping. Given the small dataset size, capping outliers was preferred to avoid data loss. Two methods were employed for capping: Winsorization (quantile method) and using the interquartile range (IQR). Winsorization was deemed more suitable due to the higher number of outliers. Z-score normalization was not applied due to the non-normalized nature of the data. Several machine learning models were trained, including XGBoost, Gradient Boosting Classifier, ETC, Random Forest and AdaBoost. Among these, XG Boost and Gradient Boosting Classifier achieved the highest accuracy. Integration of both models to create hybrid models further improved accuracy. Hyperparameter tuning and K-fold cross-validation were utilized to fine-tune these models and ensure consistent accuracy.
This research highlights how using advanced technology and machine learning can help predict heart disease more accurately. By analyzing data and using predictive tools, healthcare providers can make better decisions, leading to improved care for patients with heart problems
II. DATASET
We analyzed a dataset containing the medical records of 299 heart failure patients obtained from the UCI Machine Learning Repository. The patients consisted of 105 women and 194 men, and their ages range between 40 and 95 years old. The dataset contains 13 features, which report clinical, body, and lifestyle information, that we briefly describe here. Some features are binary: anaemia, high blood pressure, diabetes, sex, and smoking.
TABLE I FEATURES IN DATASET
|
Feature |
Explanation |
Measurement |
Range |
|
Age |
Patient’s age |
Years |
[40, ..., 95] |
|
Anaemia |
Red blood cell count reduction |
Boolean |
0, 1 |
|
High blood pressure |
Presence of hypertension in the patient |
Boolean |
0, 1 |
|
Creatinine phosphokinase |
Concentration of CPK enzyme in the blood |
Boolean |
[23, ..., 7861 |
|
Diabetes |
Presence of diabetes in the patient |
mcg/L |
0, 1 |
|
Ejection fraction |
percentage of blood leaving the heart during each contraction |
Boolean |
[14, ..., 80 |
|
Sex |
Female or Male |
Binary |
0, 1 |
|
Platelets |
Platelet count in the blood |
Kiloplatelets/mL |
[25.01, ..., 850.00] |
|
Serum creatinine |
Concentration of creatinine in the blood |
mg/dL |
[0.50, ..., 9.40] |
|
Serum sodium |
Concentration of sodium in the blood |
mEq/L |
[114, ..., 148] |
|
Smoking |
Patient's smoking status |
Boolean |
0, 1 |
|
Time |
Duration of follow- up period |
Days |
[4, ...,285] |
|
(Target) Death Event |
Patient's mortality status during the follow-up period |
Boolean |
0, 1 |
Feature extraction using Random Forest involves training a Random Forest model on a dataset containing various features and a target variable. The algorithm calculates the importance of each feature by assessing its contribution to reducing impurity in the decision trees. Features are then ranked based on their importance scores, with higher scores indicating greater predictive value. By selecting the most important features, practitioners can focus on the key predictors for improved model performance and interpretability.
An examination of data encompassing 299 patients diagnosed with heart failure, obtained in 2015, reveals that focusing solely on serum creatinine and ejection fraction is adequate for forecasting patient survival based on medical records. Intriguingly, when these two features are exclusively considered, predictions are notably more precise than when utilizing the entire array of features originally present in the dataset. This suggests that these specific indicators hold significant predictive power in determining heart failure patient outcomes, potentially simplifying predictive modelling efforts.

III. LITERATURE REVIEW
Heart failure prediction using machine learn techniques has garnered significant attention in recent years, with numerous studies highlighting the importance of specific input features in enhancing predictive accuracy. For instance, in a study by Jing Wang[1], the analysis of feature importance revealed the pivotal role of 'time' and 'serum creatinine' in determining the predictive accuracy of the model. These findings underscore the significance of identifying key features for accurate prediction.
Moreover, machine learning models and SHAP (Shapley Additive explanations) plots have emerged as valuable tools for providing clear explanations of predicted risks in disease severity assessment [2]. By elucidating the factors involved in predicting patient outcomes, clinicians can gain deeper insights into the decision-making process and tailor treatment strategies accordingly. The utilization of neural networks has shown promise in heart failure prediction, with studies highlighting their efficiency in classifying datasets without extensive pre-processing [3]. Additionally, advancements in feature selection methods, such as the comprehensive analysis of information gain, aim to identify the most informative features for predictive modeling [4]. These approaches contribute to improving model performance.
Incorporating novel kernel functions and innovative classification approaches further enhances the predictive capabilities of heart failure prediction models [5]. For instance, integrating the SMOTE (Synthetic Minority Over-sampling Technique) technique and Random Forest algorithm has resulted in accurate survival predictions while effectively handling imbalanced data [6]. Recent advancements in deep learning, including the use of Generative Adversarial Networks for heart disease diagnosis, signify notable progress in the field [7]. Machine learning and deep learning algorithms, particularly Artificial Neural Networks (ANN), have demonstrated effective predictive capability-es for cardiovascular disease prediction [8]. Techniques such as SMOTE-ENN for data balancing and feature selection significantly improve prediction accuracy in this context.
Traditional biostatistics analyses have also affirmed the importance of feature ranking carried out via machine learning [9]. Employing techniques such as Recursive Feature Elimination (RFE) with Random Forest and Logistic Regression has led to improved model performance [10]. Notably, studies have found that utilizing a subset of features yields better performance compared to using all available features [10]. The ROC curve visually illustrates the trade-off to between the true positive rate and the false positive rate for a binary classification model across different classification thresholds, aiding in model evaluation and comparison[11]. The test outcomes reveal a True Positive Rate (TP Rate) of 0.943 in the PCR Area, along with an ROC Area of 0.976 Class.
These results underscore the efficacy of the Random Forest algorithm in accurately predicting the survival of individuals with heart failure[12]. The emphasis lies in addressing the mortality rate by assessing the variables contributing to heart failure and implementing preventive measures based on the identified factors[13]. In particular, the K-Nearest Neighbors (KNN) algorithm has shown notable advancement-s in accuracy compared to prior versions, reaching impressive levels of accuracy [14]. These findings highlight the potential of machine learning algorithms in advancing heart failure prediction models. The diminished significance of certain features along with the existence of null values may be accountable for the underwhelming classification accuracy observed within the dataset[16]. Moving forward, research efforts should focus on confirming the reliability and applicability of the results, while also improving the interpretability of the algorithm-generated clusters. This will contribute to better understanding the findings and assist in making informed decisions based on the study's discoveries.[20-21]. Despite challenges, machine learning for heart disease prediction is rapidly evolving, promising enhanced accuracy and efficiency with technological advancements and increased data accessibility [18]. Overall, the literature review underscores the importance of feature selection, model interpretation, and algorithmic advancements in improving the accuracy and efficiency of heart failure prediction model.
IV. PROPOSED METHODOLOGY
After thoroughly examining various research papers, we've chosen specific models for our study. These selections were made based on their proven effectiveness, alignment with our research objectives, and relevance to our study's context. By utilizing these carefully chosen models, we aim to address our research questions effectively and contribute meaningfully to the existing body of knowledge in the field.
A. SMOTE(Synthetic Minority Over-sampling Technique)
In a project focused on predicting heart failure using machine learning, SMOTE (Synthetic Minority Over-sampling Technique) is crucial for handling imbalanced datasets where instances of heart failure are fewer than non-heart failure cases. By creating artificial samples of the minority class, SMOTE evens out the dataset, resulting in improved accuracy and prediction performance of the model. This process enables the model to effectively differentiate between heart failure and non-heart failure cases, ultimately leading to more reliable predictions. It is important to conduct thorough evaluation using accuracy and prediction metrics after applying SMOTE to ensure the model's accuracy and effectiveness in predicting heart failure.
B. Winsorization
Winsorization is a technique used to handle outliers in a dataset by substituting extreme values with less extreme ones. Outliers, which are data points that deviate significantly from the rest, have the potential to distort analyses or machine learning models. Thresholds, typically determined by percentiles or standard deviations, are established to define the range within which replacement occurs. Data points that fall beyond these thresholds are replaced, thereby reducing their impact without eliminating them entirely. This approach ensures that the data distribution is preserved, which is crucial for maintaining the integrity of the dataset. In the context of machine learning, Winsorization is employed prior to training to enhance the robustness and generalization of the model by reducing its sensitivity to outliers..
C. Random Forest
Random Forest is a machine learning algorithm that builds multiple decision trees and combines their predictions to make accurate and robust forecasts. By using random subsets of data and features, it reduces overfitting and improves generalization. Various fields use this method for classification and regression tasks.
The hyperparameters obtained following the model tuning process with GridSearchCV are as follows:
n_estimators=150, max_depth=10, min_samples_split=10, min_samples_leaf=1, max_features="auto", random_state=42
D. Gradient Boosting Classifier
Gradient Boosting Classifier improves predictive performance by combining weak learners, like decision trees, sequentially. Each new learner corrects the errors of previous ones using gradient descent optimization of a loss function. This iterative process enhances predictive accuracy, resulting in a strong ensemble model for accurate predictions.
The hyperparameters obtained following the model tuning process with GridSearchCV are as follows: n_estimators=100, learning_rate=0.2, max_depth=5, min_samples_leaf=2, min_samples_split=10, subsample=0.6, random_state=42
E. XGBoost
XGBoost is a fast and scalable machine learning algorithm used for regression and classification tasks. It sequentially adds weak learners, typically decision trees, to an ensemble using gradient descent optimization. XG Boost achieves high performance due to regularization, which prevents overfitting.
The hyperparameters obtained following the model tuning process with GridSearchCV are as follows: n_estimators=150, learning_rate=0.1, max_depth=5, subsample=0.8, colsample_bytree=0.8, random_state=42
.F. AdaBoost
AdaBoost combines weak learners like decision trees to form a strong learner. It adjusts instance weights based on previous performance, focusing more on misclassified instances in subsequent iterations. The final model combines all weak learners, resulting in a robust predictor effective for complex datasets.
The hyperparameters obtained following the model tuning process with GridSearchCV are as follows: learning_rate=0.1, n_estimators=100
G. Extra Tree Classifier (ETC)
Extra Trees Classifier builds multiple decision trees in a randomized way, increasing diversity and reducing overfitting. It further randomizes split points and thresholds, lowering variance while keeping bias low. Trees are built using random feature subsets and splitting points, and predictions are averaged across all trees. Especially when dealing with high-dimensional and noisy data, it is robust, efficient, and effective.
The hyperparameters obtained following the model tuning process with GridSearchCV are as follows: min_samples_leaf=2, min_samples_split=5, n_estimators=200
H. Hybrid model
Hybrid model in machine learning amalgamate the strengths of various techniques, such as decision trees and neural networks or supervised and unsupervised learning. By integrating these diverse approaches, they effectively handle complex data, resulting in enhanced predictive accuracy across diverse tasks and domains. In our approach, we incorported a Voting Classifier consisting of Extra Trees Classifier(ETC), Random Forest Classifier(RFC) and XGBoost models to further leverage the advantages of ensemble learning.
I. GridSearchCV
GridSearchCV is a method used in machine learning to optimize hyperparameters by exhaustively searching through a predefined grid of settings. It evaluates each combination using cross-validation, training the model on one subset of data and assessing its performance on another. The goal is to identify the hyperparameter combination that maximizes performance based on a specified metric. Although this approach is valuable for improving model effectiveness, particularly in Python's scikit-learn library, it can be computationally demanding, especially for complex models or large search spaces. Nevertheless, GridSearchCV provides a systematic and effective approach to hyperparameter tuning.
J. K-fold cross-validation
K-fold cross-validation is a vital technique in evaluating machine learning models, where the dataset is split into k equal-sized folds. In each iteration, one fold acts as the validation set, while the remaining k-1 folds are for training. This cycle repeats k times, ensuring each data point contributes to both training and validation exactly once. Maximizing data use, it offers a robust estimate of model performance. Post-iteration, metrics such as accuracy or F1-score are computed on the validation set, with the average performance across all k folds calculated. This method aids in detecting overfitting and selecting the most consistent- performing model across various validation sets, proving especially valuable with limited datasets or when precise performance assessment is crucial in model evaluation and optimization.
num_folds = 5
kf = KFold (n_splits=num_folds, shuffle=True, random_state=42)
V. IMPLEMENTATION
In the Implementation section of our research paper, we embarked on a systematic journey to analyze and model a numerical dataset, employing various techniques to ensure robustness and accuracy in our findings.

Our first step involved acquiring the dataset and conducting preliminary checks for missing values. Fortunately, the dataset exhibited completeness, sparing us the need for imputation techniques. However, we anticipated potential null values in future datasets and prepared to address them through mean imputation, ensuring data integrity. Recognizing the imbalance in our target variable, we employed the Synthetic Minority Over-sampling Technique (SMOTE) to rectify class imbalance. By synthesizing new instances of the minority class, SMOTE effectively balanced the dataset, enhancing the representativeness of our training data. With a balanced dataset in hand, we proceeded to extract significant features crucial for predictive modeling. Leveraging ensemble learning methods such as Random Forest (RF) Classifier and Extra Trees Classifier (ETC), we identified and retained the most influential features. These models, known for their robustness and ability to handle complex datasets, provided invaluable insights into feature importance.
Subsequently, we delved into outlier analysis, a critical preprocessing step to ensure model robustness. Despite the challenges posed by non-normalized data, we explored various outlier detection methods, including the Z-score method, trimming, and capping. Ultimately, we opted for capping using a quantile-based approach, effectively mitigating the impact of outliers on our models.
Having preprocessed the dataset, we partitioned it into training and testing subsets to facilitate model training and evaluation. For optimal model performance, hyperparameter tuning played a crucial role. Here, we utilized the GridSearchCV technique to systematically search through a specified parameter grid and identify the optimal combination of hyperparameters. This exhaustive search process enhanced our model’s performance and generalization ability, ensuring their efficacy across various tasks and domains. We employed k-fold cross-validation with a specific choice of k=10 folds. This approach allowed us to train and evaluate our models on diverse subsets of the data, thus providing a more reliable estimate of their performance and generalization ability. Throughout our analysis, accuracy served as the primary evaluation metric, providing a straightforward measure of predictive capability.

The crux of our implementation lay in model building and hyperparameter tuning, where we explored the efficacy of six distinct machine learning algorithms. Among these, XGBoost, ETC, and Gradient Boosting Classifier emerged as formidable contenders, known for their superior performance in predictive tasks. Additionally, we employed traditional models such as RF Classifier and ADA boost, alongside a novel hybrid model combining RF, XGBoost and ETC using a Voting Classifier ensemble approach.
Throughout the implementation process, we prioritized model validation and evaluation, leveraging techniques such as K-fold cross-validation to ensure the reliability and consistency of our model performance estimates.

VI. RESULT
Our project, built with the Streamlit framework in Python, has two main pages:
Prediction Page: Users can predict heart failure by entering relevant parameters. The model then generates predictions based on the input data.
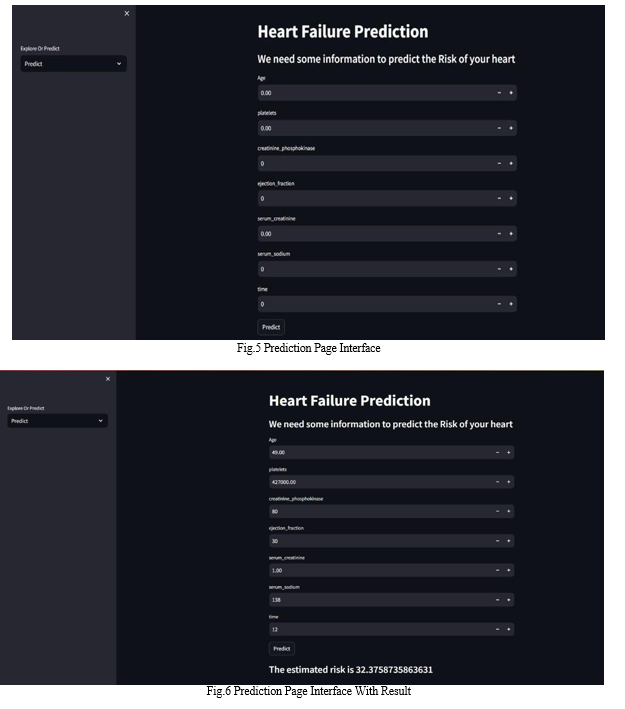

VII. FUTURE WORK
This research has the potential to enhance healthcare by providing a valuable tool for predicting heart failure survival, aiding healthcare providers in identifying patients at risk. By understanding which patients are likely to survive, physicians can prioritize major risk factors and tailor treatment accordingly. Future research could explore using combinations of different machine learning models to leverage their individual strengths. Additionally, there's room for improvement in feature selection techniques to enhance the performance of machine learning models further.
Our project model possesses the capability to seamlessly integrate with ECG (Electrocardiogram) machines. This integration empowers the model to instantly predict the estimated risk level of the patient at the moment of testing. By directly interfacing with ECG machines, our system can analyze real-time data and provide timely insights, aiding healthcare professionals in making informed decisions regarding patient care and management.
VIII. ACKNOWLEDGMENTS
- We extend our gratitude to the UCI portal for providing the dataset, which played a pivotal role in our project.
- Additionally, we acknowledge the Streamlit library in Python for significantly simplifying the development and deployment of our project.
- The teamwork, guidance, and collaborative efforts throughout this project have been instrumental in refining our ideas, overcoming challenges, and achieving our objectives. We extend our heartfelt thanks to Dr.N. Sri Hari our project coordinator and Mr. M. JeevanBabu our project mentor, for their invaluable advice, encouragement, and insightful feedback throughout the project duration. Their expertise and support have played a pivotal role in shaping our strategy and navigating hurdles.
Conclusion
In summary, our study demonstrates the effectiveness of machine learning algorithms in predicting the survival of heart patients. Through the utilization of various techniques including XGBoost, Extra Trees Classifier (ETC), AdaBoost, Random Forest-(RF), Gradient Boosting Classifier(G-BC), and a hybrid model incorp-orating RF, XGBoost, and ETC, coupled with the Synthetic Minority Over-sampling Technique (SMOTE) to address class imbalance, we successfully identified key Predictive features such as age, anaemia, and others.
References
[1] Jing Wang 2021 J. Phys.: Conf. Ser. 2031 01206 Heart Failure prediction with Machine Learning: A comparative study Date of publication: - 2021 [2] Ke Wang, Jing Tian, Chu Zheng, Hong Yang, Jia Ren1 Chenhao Li1, Qinghua Han, Yanbo Zhang, Improving Risk Identification of Adverse Outcomes in Chronic Heart Failure Using SMOTE +ENN and Machine Learning [3] Muhammad Waqar, Hassan Dawood, Hussain Dawood, Nadeem Majeed, Ameen Banjar and Riad Alharbey5An Efficient SMOTE-Based Deep Learning Model for Heart Attack Prediction Volume 2021, Article ID 6621622, 12 pages [4] Jian Yang and Jinhan GuanA Heart Disease Prediction Model Based on Feature Optimization and Smote, Boost Algorithm. Date of publication: 2 October 2022 [5] Rohit Baxani, Maniroja Edinburgh Heart Disease Prediction Using Machine Learning Algorithms Logistic Regression, Support Vector Machine and Random Forest Classification Techniques [6] Mohammed Mafaz Survival analysis of heart failure using Machine Learning on an imbalanced dataset Date of publication: 2021.Available on: https://www.ijariit.com Volume 7, Issue 4 - V7I4-1655 [7] Md Manjurul Ahsan, Zahed Siddique MACHINE LEARNING-BASED HEART DISEASE DIAGNOSIS: A SYSTEMATIC LITERATURE REVIEW Date of publication: DECEMBER 14, 2021 [8] Avvaru R V Naga Suneetha, Dr. T. Mahalngam Cardiovascular Disease Prediction Using ML and DL Approaches Date of publication: 12 September 2022 [9] Davide Chicco and Giuseppe Jurman Machine learning can predict survival of patients with heart failure from serum creatinine and ejection fraction alone. Date of publication: 2020 [10] Shivanshi Shukla, Usha Chouhan, Sunil Kumar Suryawanshi, Jyoti Kant Choudhari A comparative study by using machine Learning classifiers to enhance classification and prediction heart failure disease. Date of publication:22 Aug 2022 [11] Nabaouia Louridi, Samira Douzi, Bouabid El Ouahid Machine learning-based identification of patients with a cardiovascular defect. Date of Publication:2021 [12] Sri Rahayu, Jajang Jaya Purnama, Achmad Baroqah Pohan, Fitra Septia Nugraha, Siti Nurdiani, Sri Hadiant PREDICTION OF SURVIVAL OF HEART FAILURE PATIENT USING RANDOM FOREST. Date of publication:2 sep 2020 [13] Sudipta Priyadarshinee, Madhumita Panda CARDIAC DISEASE PREDICTION USING SMOTE AND MACHINE LEARNING CLASSIFIERS Publication:2022 [14] Harshit Jindal1, Sarthak Agrawal1, Rishabh Khera1, Rachna Jain2 and Preeti Nagrath2Heart disease prediction using Machine Learning algorithm Publication:2021 [15] Prasanta Kumar Sahoo, Pravalika Jeripothula HEART FAILUR PREDICTION USING MACHINE LEARNING TECHNIQUES [16] Md. Julker Nayeem, Sohel Rana, and Md. Rabiul Islam Prediction of Heart Disease Using Machine Learning Algorithms. Date of publication:30 Nov 2022. [17] Maria Teresa Garcia-Ordas, Martin Bayon-Gutierrez, Carmen Benavides, Jose Aveleira-Mata, Jose Alberto Benitez-Andrades Heart Disease risk Prediction using deep Learning techniques with feature Augmentation. Date of Publication:6 Feb 2023 [18] Kalpesh Joshi, Shubham Patil, Sagar Patil, Sahil A. Patil, Shantanu Patil, Saniya Patil Heart Disease Prediction Using Machine Learning. Date of Publication: June 2023 [19] Rishabh Magar, Rohan Memane, Suraj Raut, Prof. V. S. Rupnar Heart Disease Prediction Using Machine Learning. Date of Publication: June 2020.Volume:7 [20] Chintan M. Bhatt, Parth Patel, Tarang Ghetia, Pier Luigi Mazzeo Effective Heart Disease Prediction Using Machine Learning Techniques. Date of Publication: June 2023. Volume:16 [21] Jeevan Babu Maddala,M.Vanaja,P.Satya,N.Harika,N.Dinesh, CHRONIC KIDNEY DISEASE PREDICTION.Date of Publication:December 2022.Volume:11
Copyright
Copyright © 2024 Jeevan Babu Maddala, Bhargav Reddy Modugulla, Sahithi Amulya Pulusu, Sanjay Mannepalli, Praveen prakash Pamidimalla, Rukhiya Khanam. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET59236
Publish Date : 2024-03-20
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

