Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
INSILICO Analysis of CADD Approach for Alzheimer Disease Caused by Heavy Metals in Humans
Authors: Uma Kumari, Taneya Gupta, Chaitanya Gupta, Navjot Virk
DOI Link: https://doi.org/10.22214/ijraset.2023.54875
Certificate: View Certificate
Abstract
Alzheimer\'s disease (AD) stands as a well-studied neurodegenerative disorder and remains the foremost cause of dementia. Its nomenclature pays homage to the groundbreaking work of Alois Alzheimer, a pivotal figure in elucidating its characteristics. AD is characterized by a progressive decline in cognitive function, accompanied by impairments in daily activities and noticeable changes in behavior. Alois Alzheimer\'s seminal research significantly contributed to the identification of amyloid plaques and the decline in neurons within the brain, laying the foundation for our contemporary comprehension of the disease.Alzheimer\'s disease (AD) stands as a well-studied neurodegenerative disorder and remains the foremost cause of dementia. Its nomenclature pays homage to the groundbreaking work of Alois Alzheimer, a pivotal figure in elucidating its characteristics. AD is characterized by a progressive decline in cognitive function, accompanied by impairments in daily activities and noticeable changes in behavior. Alois Alzheimer\'s seminal research significantly contributed to the identification of amyloid plaques and the decline in neurons within the brain, laying the foundation for our contemporary comprehension of the disease.
Introduction
I. INTRODUCTION
Alzheimer's disease (AD) remains extensively researched neurodegenerative disorder and stands as the primary manifestation of dementia. The nomenclature itself is a tribute to the distinguished contributions of Alois Alzheimer, the esteemed German psychiatrist, who played a pivotal role in its early comprehension. AD exhibits a progressive deterioration of cognitive faculties, posing substantial obstacles to the execution of everyday tasks and marked changes in behavioral patterns over time [1, 2].His groundbreaking research centered on the meticulous examination of his first patient's brain, yielding invaluable revelations about the ailment. During the initial assessment, he made profound observations that would lay the foundation for our current comprehension of the disorder we now recognize as Alzheimer's disease. Notably, he identified the existence of amyloid plaques, anomalous protein deposits, alongside a remarkable decline in the neuronal population. This patient exhibited a constellation of symptoms, including memory impairment, personality alterations, ultimately succumbing to the disease's progression [3].Alois Alzheimer astutely acknowledged the gravity of this affliction and its particular influence on the cerebral cortex, the area of the brain crucial for advanced cognitive processes. These significant discoveries played a pivotal role in establishing the groundwork for subsequent research and clinical examinations regarding the process and advancement of Alzheimer's disease. The formal introduction of the term "Alzheimer's disease" was credited to Emil Kraepelin in the 8th edition of his esteemed psychiatry handbook, solidifying its position within the realm of medical literature [4].Alzheimer's disease stands apart due to the detection of neurofibrillary tangles and extracellular amyloid plaques within the brainof human. These pathogenic elements entail hyperphosphorylate τ-protein, actively contributing to the gradual decline of cognitive abilities characteristic of this condition. Remarkably, the worldwide scale of Alzheimer's disease bears profound implications, with approx 47 million folks currently grappling with dementia's impact across the globe [5].Anticipated by experts, there is an imminent surge in these figures, with projections indicating that by the year 2050, the dementia-affected population will surpass 131 million individuals, signifying a remarkable threefold increment [6]. These foresights underscore the pressing necessity for persistent research and the formulation of efficacious approaches for diagnosis and treatment addressing the escalating impact of AD [7].
The primary aim of this study was to explore the three-dimensional structural attributes, domains, and functionalities of mutated Alzheimer's disease proteins, along with the characteristics of associated ligand preparations.
To investigate the interaction between the mutant gene responsible for Alzheimer's disease in humans and its corresponding ligand, a molecular docking analysis will be conducted. Additionally, the research will entail in silico analysis and computer-aided medication development for Alzheimer's disease, along with predicting the metal configuration and its position and orientation within the binding affinity.
II. MATERIAL AND METHODS
At the core of bioinformatics lies a fundamental instrument, a computer program designed to emulate the intricate functions of biological information systems, thereby offering predictions and novel perspectives within the realm of biology. This program conducts computations that furnish biologists with vital insights into DNA sequences, protein structures, gene regulatory mechanisms, and various other biological entities and processes. The bedrock of this field resides in the application of computer software, a principle that underpins the study. To acquire sequences, the investigation made use of the esteemed National Center for Biotechnology Information (NCBI).NCBI stands as an abundant repository of databases dedicated to biotechnology and biomedicine, offering a diverse array of bioinformatics tools and services. Prominent examples of these resources include GenBank, a comprehensive collection of DNA sequences, and PubMed, a renowned platform for biomedical literature. Additionally, the study also made use of the vital resource known as the Protein Data Bank (PDB), a crucial archive housing essential structural data pertaining to biological macromolecules like proteins, nucleic acids, and intricate assemblies.For molecular docking investigations, the research employed docking software tools such as RasMol, PyMol, and AutoDockVina. RasMol serves as an open-source platform specifically designed for the display and analysis of biological molecules, with a particular focus on proteins, nucleic acids, and small molecules. It offers an extensive range of functionalities, enabling selection, coloring, labeling, and rendering of the molecule, along with an interactive environment to visualize and manipulate 3D molecular structures. PyMOL, created by Warren Lyford DeLano and initially distributed by DeLano Scientific LLC, is a freely available software system catering to the visualization of macromolecules. On the other hand, AutoDockVina, widely utilized open-source software, is known for its speed and efficiency in molecular docking procedures.
III. RESULTS
Information about the target, such as the PDB ID (2MGT), was retrieved from the NCBI database and used in Computer-Aided Drug Design (CADD). The target's 3D structure was then obtained from the Protein Data Bank (PDB) database in PDB format. In this particular project, the target under scrutiny is denoted as 2MGT, constituting a dimer of the metal binding domain 1-16 of human amyloid beta-peptide, instigated by Zinc and bearing the H6R English mutation, which is linked to the pathogenesis of Alzheimer's disease.Next, heavy metal structures of Copper, Iron, Lead, Cadmium, and Manganese were required as ligands. To obtain these structures, PubChem database was accessed and downloaded the structures of these metals. These structures will be utilized in the later stages of the project. The process involved the selection and downloading of the necessary structures from the PubChem database. In order to study the structure of the target molecule, we utilized the RasMol software. This software allows us to read molecular coordinates from a file and interactively display the molecule in various representations. Specifically, we used RasMol to analyze the structure of target 2MGT and gain a deeper understanding of its molecular composition.After launching RasMol, the target structure was loaded into the software.Through this process, we were able to visualize the target in a variety of representations, allowing us to examine its structure from multiple perspectives.By utilizing these options, more comprehensive understanding of the structure and its features was gained.
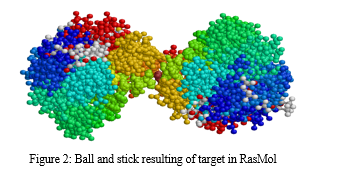
In this, the target molecule employs a diverse color scheme to differentiate between its constituent atoms. Carbon atoms are conventionally depicted in shades of gray or black, while nitrogen atoms take on a blue hue, and oxygen atoms are visually identified through a striking red color. Likewise, sulfur atoms adopt a vibrant yellow appearance, phosphorus atoms display an orange shade, and hydrogen atoms remain distinct in pristine white.
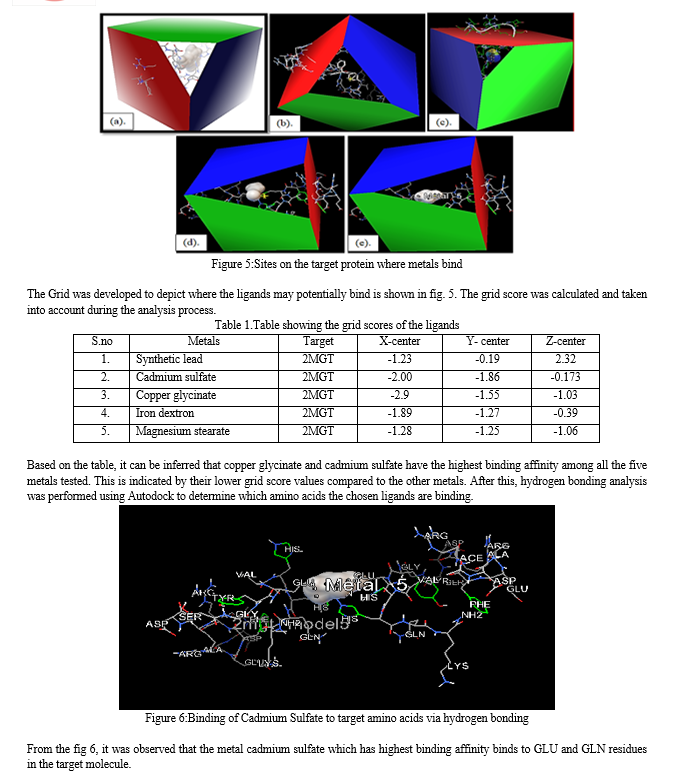
A more in-depth presentation of the atoms and bonds was seen in the representation of the structure. Through these representations, it was determined that the target molecule only had an alpha helix and did not have a beta-helix. In particular, the alpha helix was highlighted in a magenta color, which enabled us to readily differentiate it from the structure's other components.The hydrogen bonds that connect various parts of the molecule can be visualized by using Rasmol's chain form representation of the molecule.After this, heavy metal structures of copper, iron, cadmium, and magnesium, lead which were selected to act as ligands were downloaded from PubChem. PyMol software was utilized to observe the interactions between the target and ligands, which provided valuable insight into the binding location of themetals with target. The results gave the broad overview of how and where the metals will bind in thetarget protein.The analysis revealed that synthetic lead (a), cadmium sulfate (b), copper glycinate (c), iron dextron (d), and magnesium stearate (e) bind to the target protein. Additionally, the analysis revealed the particular residues that participated in the binding process. Specifically, the GLU and HIS residues of the target protein were involved in the binding of magnesium stearate, while the binding of cadmium sulfate was linked with the HIS residue of the target protein.The next step involved preparing the target and the ligands. For the target preparation, model 5 of the target protein was selected for docking. The five different metals were then prepared and docked with the target protein using Command Prompt. The objective was to identify the binding sites and determine their respective docking scores. Finally, the results of the docking process were visualized for analysis.
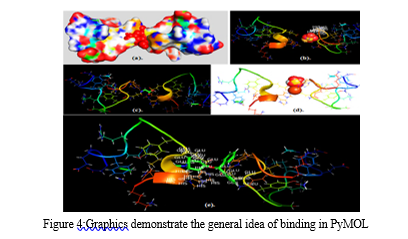
IV. DISCUSSION
Alzheimer's disease (AD) is a prevalent form of dementia, characterized by a progressive degeneration of neurological function over time. This neurodegenerative condition leads to a decline in cognitive abilities, loss of independence in daily activities, and noticeable behavioral changes. Understanding the factors and stages of AD progression is crucial for developing effective therapies and interventions. In this study, we focused on investigating the role of the zinc-induced dimer of the metal-binding domain 1-16 of human amyloid beta-peptide (2MGT) as the target protein, which has been linked to Alzheimer's disease. Existing research suggests that structural changes in the Aβ peptide result in the formation of hazardous soluble dimers and non-soluble complexes from a single molecule, which is a characteristic feature of AD.The N-terminal domain Aβ (1-16) has been identified as responsible for facilitating the binding of zinc ions with Aβ, making it a significant factor in the progression of Alzheimer's disease. It is essential to note that metals are naturally occurring elements found in both biological systems and the environment. While they are necessary for proper physiological functioning, imbalances or disruptions in the regulation of certain metal ions, critical for neurological processes, can lead to neurotoxicity and damage to the nervous system. Thus, maintaining appropriate levels of these metal ions is essential in preventing such damage.To explore the effects of various metals on the target protein and their potential role in the development of Alzheimer's disease, the study selected five heavy metals, including lead, copper, cadmium, manganese, and iron, as ligands for the target protein.To initiate the investigation, the we employed RasMOL and PyMol to analyze the structure of the target protein and the ligands, allowing them to gain valuable insights from multiple perspectives. By visualizing the various atoms in the target structure, such as carbon, nitrogen, oxygen, sulfur, phosphorus, and hydrogen, they were able to identify the crucial amino acids present.Furthermore, we observed that the target protein solely consisted of an alpha helix and lacked a beta helix. This structural information played a pivotal role in further examinations.To gain an overview of the binding sites of the heavy metal ligands, the study utilized PyMol to explore the interactions between the target protein and these metals after partially preparing the ligands.The results indicated that lead, cadmium sulfate, copper glycinate, iron dextron, and magnesium stearate were all capable of binding to the target protein, with the GLU and HIS residues playing a particularly significant role in the binding process.For a more in-depth analysis of the binding modes, binding energies, and protein-ligand interactions, AutoDockVina was employed. This computational tool enabled the researchers to predict potential binding locations of the ligands and calculate their binding affinity using the grid score method.The calculated grid scores revealed that cadmium sulfate exhibited the highest binding affinity, indicating its strong potential for interaction with the target protein. Lower grid scores in ligands suggest higher binding energies, indicating a stronger affinity for the protein. Conversely, higher scores suggest weaker predicted binding affinities.From the hydrogen bonding analysis, it was found that the metal cadmium sulfate, with the highest binding affinity, formed interactions with the GLU and GLN residues in the target molecule.Overall, this study contributes valuable insights into the molecular interactions between heavy metal ions and the target protein associated with Alzheimer's disease. The findings underscore the potential role of metal ions in AD pathogenesis, suggesting that maintaining proper metal ion levels could be a relevant therapeutic strategy.However, it is crucial to acknowledge the limitations of this research. The study relied on computational simulations, and while they provide valuable predictions, further experimental validations are essential to confirm the results. Moreover, the complexity of Alzheimer's disease requires a multidisciplinary approach, and additional studies are needed to comprehensively understand the intricate mechanisms involved in its progression.
Conclusion
In conclusion, this study represents a significant step towards unraveling the molecular underpinnings of Alzheimer\'s disease and the potential involvement of heavy metal ions in its pathogenesis. The findings contribute to our understanding of AD and open up promising avenues for future research and therapeutic developments. The ongoing efforts to explore the complex interactions between metal ions and Alzheimer\'s-related proteins hold the potential to shape novel and effective strategies to combat this devastating neurodegenerative conditions. Further exploration of the role of heavy metal ions in the aggregation and neurotoxicity of amyloid beta-peptide could lead to novel therapeutic approaches and preventive strategies for Alzheimer\'s disease.
References
[1] De-Paula VJ, Radanovic M, Diniz BS, Forlenza OV. Alzheimer\'s disease. SubcellBiochem. 2012;65:329-352. [2] Citron M. Alzheimer\'s disease: strategies for disease modification. Nat Rev Drug Discov. 2010;9(5):387-398. [3] Hippius H, Neundörfer G. The discovery of Alzheimer\'s disease. Dialogues ClinNeurosci. 2003;5(1):101-108. [4] Cipriani G, Dolciotti C, Picchi L, Bonuccelli U. Alzheimer and his disease: a brief history. Neurol Sci. 2011;32(2):275-279. [5] Knopman DS, Amieva H, Petersen RC, et al. Alzheimer disease. Nat Rev Dis Primers. 2021;7(1):33. [6] DeTure MA, Dickson DW. The neuropathological diagnosis of Alzheimer\'s disease. MolNeurodegener. 2019 Aug 2;14(1):32. [7] Tiwari S, Atluri V, Kaushik A, Yndart A, Nair M. Alzheimer\'s disease: pathogenesis, diagnostics, and therapeutics. Int J Nanomedicine. 2019;14:5541-5554. [8] Colomina MT, Peris-Sampedro F. Aluminum and Alzheimer\'s Disease. AdvNeurobiol. 2017;18:183-197. [9] Huat TJ, Camats-Perna J, Newcombe EA, Valmas N, Kitazawa M, Medeiros R. Metal Toxicity Links to Alzheimer\'s Disease and Neuroinflammation. J Mol Biol. 2019;431(9):1843-1868. [10] Goedert M. NEURODEGENERATION. Alzheimer\'s and Parkinson\'s diseases: The prion concept in relation to assembled A?, tau, and ?-synuclein. Science. 2015;349(6248):1255555. [11] Chen JX, Yan SS. Role of mitochondrial amyloid-beta in Alzheimer\'s disease. J Alzheimers Dis. 2010;20Suppl 2:S569-S578. [12] Crews L, Masliah E. Molecular mechanisms of neurodegeneration in Alzheimer\'s disease. Hum Mol Genet. 2010;19(R1):R12-R20. [13] O\'Brien RJ, Wong PC. Amyloid precursor protein processing and Alzheimer\'s disease. Annu Rev Neurosci. 2011;34:185-204. [14] Selkoe DJ. Cell biology of the amyloid beta-protein precursor and the mechanism of Alzheimer\'s disease. Annu Rev Cell Biol. 1994;10:373-403. [15] Hefter D, Kaiser M, Weyer SW, et al. Amyloid Precursor Protein Protects Neuronal Network Function after Hypoxia via Control of Voltage-Gated Calcium Channels. J Neurosci. 2016;36(32):8356-8371. [16] Selkoe DJ. Cell biology of protein misfolding: the examples of Alzheimer\'s and Parkinson\'s diseases. Nat Cell Biol. 2004;6(11):1054-1061. [17] Kimberly WT, Zheng JB, Guénette SY, Selkoe DJ. The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J Biol Chem. 2001;276(43):40288-40292. [18] Bergmans BA, De Strooper B. gamma-secretases: from cell biology to therapeutic strategies. Lancet Neurol. 2010;9(2):215-226.
Copyright
Copyright © 2023 Uma Kumari, Taneya Gupta, Chaitanya Gupta, Navjot Virk. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET54875
Publish Date : 2023-07-20
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

