Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Liver Cancer Prediction Using Matlab
Authors: G. Srikanth, Kalva Deepika, Nayini Uday Kiran, Ramala Santhosh, Gaddam Devika
DOI Link: https://doi.org/10.22214/ijraset.2024.60033
Certificate: View Certificate
Abstract
The liver is necessary for survival and is also prone to many diseases. CT examinations can be used to plan and properly administer radiation treatments for cancers and to guide biopsies and other minimally invasive procedure. The statistical and textural information are obtained from the extracted cancer using the features like mean, standard deviation and entropy of the obtained sub bands are calculated and stored in a feature vector (in format of mat file). The extracted features are fed as input to Extreme Machine Learning classifier to identify the presence of Liver cancer disease and to classify it as Malignant or Benign stage.
Introduction
I. INTRODUCTION
A liver cancer or intracranial neoplasm occurs when abnormal cells from within the liver. There are two main types of cancers: malignant or cancerous cancers and benign cancers. Cancerous cancers can be divided into primary cancers that start within the liver, and secondary cancers that have spread from somewhere else, known as liver metastasis cancers. Diagnosis is usually by medical examination along with computed tomography or CT imaging. Liver cancers are not a very common disease, but they are among the most fatal cancers. Reported an incidence of less than 1 h in the western population but life expectancy for an individual can be one year or even less for the most aggressive liver cancers. The causes of liver cancer are still largely unknown, the only environmental risk factors which could be identified so far, are exposure to certain chemicals or ionizing radiation.
Humans are considered as the most unique creation of nature. Cancer being one of the deadliest and most widespread disease is the most fatal one. Across the globe efforts are being made to cure it and eradicate the disease. No 100% cure has yet been developed but treatment like chemotherapy and other intense radiation passing on the affected area are helpful treatments in controlling the disease. This becomes even more crucial, if there is a medical emergency. Though cancer can be detected in human body through CT images, but the intensity of CT images and also the shape complexity of organs pose a threat to accurate detection. Also, during the diagnosis and treatment phase the doctors are majorly interested in the problem area and not the entire human body. That is the reason that image segmentation comes into play. Segmentation sub divides the area of interest to provide a better and clear view of the organ or part under observation. It should be noted that, segmentation is only a pre-treatment step. The detection of cells or of structures interior to cells can be considered as an image segmentation problem within digital image analysis. Different algorithms can be employed to segment an image. The algorithm discussed in this paper is Genetic algorithm. Watershed algorithm: Multi threshold values in CT images are interpreted on the basis of grey levels. Comprehensive methods take advantage of de-noising and gradient construction. The grey level of a pixel is interpreted as its altitude. Local minimum values are set. Intuitively, the watershed of a relief corresponds to the limits of the adjacent catchment basins of the drops of water. Generic Algorithm: Using the binary strings, optimization problems are solved. This algorithm is inspired by natural evolution. Once the random variables are generated, these can be improved by iteratively applying operators, termed selection, crossover and mutation that mimic the corresponding processes of natural evolution. In fact, selection lets only the fittest individuals to be present in the next generation (iteration of the algorithm); crossover lets them exchange tracts of their DNA (corresponding substrings) to generate offspring (new solutions), while mutation randomly introduces new genes (by flipping one or more bits of a solution). Image processing is a developing and growing field in context to the medical application. Many methods have been developed and replaced with the newer methods. So, it becomes of prime importance to develop and select newer methods to suit the requirements of the current times and problem specifications. Likewise, 3D image analysis, reconstruction of the CT slices and accurate boundary detection are of prime importance. Softening of cells is a major problem in cancer, and various 3D orthogonal planes (sagittal, coronal, transverse) are acquired. Due to the shape of liver, its overlapping regions with lungs and heart and the artifacts of motion and pulsation automatic liver segmentation is a difficult process, also the CT images show grayish values of range between 90-92 out of 0255 for a normal cancer free liver, but if there is cancer then the images become darker and the range is also ambiguous. So, it is felt that it’s high time to design and implement a quick responsive and exact calculative liver segmentation method for medical image analysis, which supports to analyse the benefits and problems of liver transplantation and the treatment method of liver cancers.
CT imaging is a far better method than CT scan for the reason of being free of ionizing radiation and also gives a better image of soft tissue in terms of visualization.
II. RELATED WORKS
A. Machine Learning Approaches for Liver Cancer Prediction: A Review
This paper provides a comprehensive review of machine learning approaches used for liver cancer prediction. It discusses various techniques such as logistic regression, decision trees, support vector machines (SVM), and deep learning models applied to liver cancer prediction datasets. The review highlights the strengths and limitations of each approach and identifies current research trends and challenges in the field.
B. Feature Selection Methods for Liver Cancer Prediction Models
This paper focuses on feature selection methods tailored specifically for liver cancer prediction models. It evaluates techniques such as filter, wrapper, and embedded methods for selecting the most relevant features from large-scale liver cancer datasets. The study compares the performance of different feature selection algorithms in terms of prediction accuracy, computational efficiency, and robustness.
C. Comparative Analysis of Machine Learning Models for Liver Cancer Prediction
This paper presents a comparative analysis of various machine learning models for liver cancer prediction using MATLAB. It evaluates the performance of different algorithms, including SVM, logistic regression, random forests, and neural networks, on benchmark liver cancer datasets. The analysis includes metrics such as accuracy, sensitivity, specificity, and area under the receiver operating characteristic curve (AUC-ROC) to assess model performance comprehensively.
D. Integration of Genetic Algorithms with Machine Learning for Liver Cancer Prediction
This paper explores the integration of genetic algorithms (GAs) with machine learning techniques to optimize liver cancer prediction models. It investigates the use of GAs for feature selection, hyperparameter tuning, and model optimization to improve prediction accuracy and robustness. The study demonstrates the effectiveness of GA-based approaches in enhancing the performance of liver cancer prediction models.
E. Deep Learning Approaches for Liver Cancer Prediction Using MATLAB
This paper investigates the application of deep learning techniques, such as convolutional neural networks (CNNs) and recurrent neural networks (RNNs), for liver cancer prediction using MATLAB. It explores the use of deep learning architectures for automatic feature extraction and hierarchical representation learning from raw liver cancer data. The study evaluates the performance of deep learning models in comparison to traditional machine learning algorithms and discusses their potential advantages and limitations for liver cancer prediction tasks.
III. METHODS
A. Statistical Analysis
Machine Learning Algorithms
MATLAB offers a wide range of machine learning algorithms that can be used for liver cancer prediction. These include:
- Logistic Regression
- Decision Trees
- Support Vector Machines (SVM)
- Neural Networks: MATLAB's
B. Ensemble Methods
Deep Learning
Rule-based Systems
Feature Selection and Dimensionality Reduction
MATLAB provides methods for feature selection and dimensionality reduction to enhance model performance and reduce computational complexity.
By leveraging these different methods of implementation within MATLAB, researchers and practitioners can develop robust and accurate liver cancer prediction models tailored to specific datasets and clinical settings.
IV. IMPLEMENTATION AND RESULTS
A. Implementation
Here are the implementation steps for developing a liver cancer prediction system using MATLAB:
- Data Collection and Preprocessing: Gather a dataset containing relevant patient information such as demographics, medical history, liver function tests, and cancer diagnosis.
- Exploratory Data Analysis (EDA): Conduct exploratory data analysis to understand the distribution and relationships between different variables.
- Feature Selection and Engineering: Select relevant features based on domain knowledge, statistical analysis, or machine learning techniques.
- Model Selection: Choose appropriate machine learning algorithms for classification tasks, considering factors such as dataset size, complexity, and interpretability.
- Model Training: Split the dataset into training and testing sets to evaluate model performance.
- Hyperparameter Tuning: Tune hyperparameters of the models to optimize their performance using techniques like grid search or random search.
- Model Evaluation: Evaluate the trained models using appropriate evaluation metrics such as accuracy, precision, recall, F1-score, and area under the ROC curve.
- Deployment and Integration: Deploy the selected model into a MATLAB-based application or script for real-time prediction of liver cancer risk.
- Validation and Testing: Validate the deployed model using independent datasets or through collaboration with medical experts.
- Documentation and Reporting: Document the entire implementation process, including data preprocessing steps, model selection criteria, training procedures, and evaluation results.
By following these implementation steps, researchers and practitioners can develop a robust and accurate liver cancer prediction system using MATLAB, contributing to improved patient outcomes and healthcare decision-making. Ensure that the system has a backup power supply to prevent disruption in case of power outages.
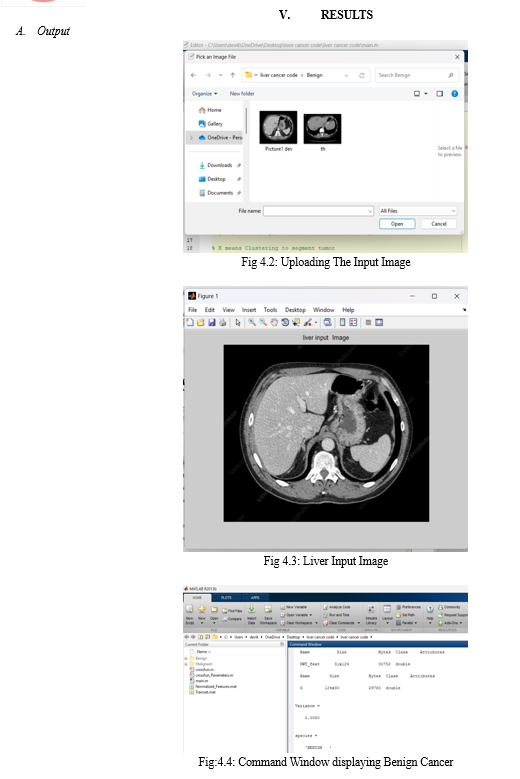
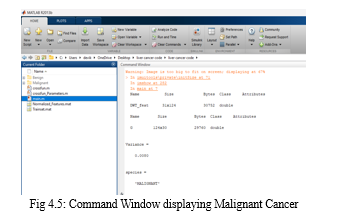
VI. FUTURE SCOPE
The future scope of the project is the FUZZY C MEAN Technology.
Some of the FCM features are:
- Future advancements in Fuzzy C-means (FCM) may focus on optimizing parameter selection and initialization strategies to improve clustering accuracy and convergence speed.
- Research could explore novel variations of FCM that incorporate adaptive learning mechanisms or incorporate constraints to better capture complex data relationships.
- Integration of FCM with deep learning frameworks or reinforcement learning techniques could unlock new capabilities for unsupervised learning tasks and pattern recognition in dynamic environments.
VII. ACKNOWLEDGEMENT
Sincere thanks of gratitude are extended to the Guide Mr. G. Srikanth”, Professor and Head, of the Department of Electronics & Communication Engineering and coordinator “Dr. Sudha Arvind”, Professor, Department of Electronics & Communication Engineering, for their guidance and support in completing the project and also to “Mr. G. Srikanth”, Professor and Head, of the Department of Electronics & Communication Engineering, and “Dr. A. Raji Reddy” Director of CMR Technical Campus for providing all the facility that was required.
Conclusion
Different CT Images were acquired from the internet, basic preprocessing technique was used, for histogram process was used and it was observed that for a few images’ segmentation was done correctly, so our future works includes Suppose if it is a mass then K- means algorithm is enough to extract it from the liver cells. If there is any noise are present in the CT image it is removed before the K-means process. Classification based segmentation segment cancer accurately and manufacture sensible results for big information set however undesirable behaviours can occur in case wherever a category is unrepresented in training data. Clustered based segmentation performs is straight forward, quick and manufacture sensible results . In spite of several dealing of problems, an atomization of livercancer segmentation using combination of threshold based and classification with SVM overcame the problems and gives effective and accurate results for livercancer detection.
References
[1] Zhang, Y.,L Zhang, Y., L. Wu, and S. Wang, “CTliver image classification by an improved artificial bee colony algorithm,” Progress In Electromagnetics Research, Vol. 116, 65–79, 2011. [2] Mohsin, S. A., N. M. Sheikh, and U. Saeed, “CT induced heating of deep liver stimulation leads: Effect of the air-tissue interface,” Progress In Electromagnetics Research, Vol. 83, 81–91, 2008. [3] Golestanirad, L., A. P. Izquierdo, S. J. Graham, J. R. Mosig, and C.Pollo, “Effect of realistic modeling of deep liver stimulation on the prediction of volume of activated tissue,” Progress In Electromagnetics Research, Vol. 126, 1–16, 2012. [4] Mohsin, S. A., “Concentration of the specific absorption rate around deep liver stimulation electrodes during CT,” Progress In Electromagnetics Research, Vol. 121, 469–484, 2011. [5] Oikonomou, A., I. S. Karanasiou, and N. K. Uzunoglu,“Phasedarray near field radiometry for liver intracranial applications,” Progress In Electromagnetics Research, Vol. 109, 345–360, 2010. [6] Scapaticci, R., L. Di Donato, I. Catapano, and L. Crocco, “A feasibility study on microwave imaging for liver stroke monitoring,” Progress In Electromagnetics Research B, Vol. 40, 305–324, 2012. [7] Asimakis, N. P., I. S. Karanasiou, P. K. Gkonis, and N. K. Uzunoglu, “Theoretical analysis of a passive acoustic liver monitoring system,” Progress In Electromagnetics Research B, Vol. 23, 165–180, 2010. [8] Chaturvedi, C. M., V. P. Singh, P. Singh, P. Basu, M. Singaravel, R. K. Shukla, A. Dhawan, A. K. Pati, R. K. Gangwar, and S. P. Singh.“2.45GHz (CW) microwave irradiation alters circadian organization, spatial memory, DNA structure in the liver cells and blood cell counts of male mice, mus musculus,” Progress In Electromagnetics Research B, Vol. 29, 23–42, 2011. [9] Emin Tagluk, M., M. Akin, and N. Sezgin, “Classification of sleep apnea by using wavelet transform and artificial neural networks,” Expert Systems with Applications, Vol. 37, No. 2, 1600–1607, 2010
Copyright
Copyright © 2024 G. Srikanth, Kalva Deepika, Nayini Uday Kiran, Ramala Santhosh, Gaddam Devika. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET60033
Publish Date : 2024-04-08
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

