Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Machine Learning-Based Diagnosis and Genetic Analysis of C9ORF72-Associated Amyotrophic Lateral Sclerosis (ALS)
Authors: Hans Elkan Sam C
DOI Link: https://doi.org/10.22214/ijraset.2023.56069
Certificate: View Certificate
Abstract
Amyotrophic Lateral Sclerosis (ALS), commonly known as Lou Gehrig\'s disease, is a devastating neurodegenerative disorder characterized by motor neuron degeneration. This research article explores the use of machine learning algorithms to diagnose ALS by analyzing mutations and insertions/deletions (indels) in the C9orf72 gene. The study utilizes a dataset to predict three stages of ALS: normal, intermediate risk, and full mutation. Three machine learning algorithms, Random Forest, SVM, and k-NN, were employed for diagnosis. Results indicate high accuracy in identifying ALS stages, with Random Forest and SVM achieving perfect accuracy when considering mutations in the Allele2. The findings demonstrate the potential of machine learning in early ALS diagnosis, offering insights for improved patient care and genetic understanding of the disease.
Introduction
I. INTRODUCTION
Amyotrophic Lateral Sclerosis (ALS), often referred to as Lou Gehrig's disease, is a rare and devastating neurological disorder that has intrigued scientists and clinicians for decades. ALS most commonly affects people of any racial or ethnic group between the ages of 40 and 70, although it can occur at a younger age. The disease primarily targets motor neurons, specialized nerve cells responsible for transmitting signals from the brain and spinal cord to muscles throughout the body. As these motor neurons progressively degenerate and perish, patients experience a range of debilitating symptoms. Early signs include muscle weakness, muscle atrophy, and muscle fasciculation or twitching, often accompanied by difficulties in speaking, swallowing, and breathing. Eventually, ALS leads to complete paralysis and, sadly, death, typically within 2 to 5 years of diagnosis.
The exact cause of ALS remains largely unknown despite extensive research. ALS can be categorized into two main types: sporadic and familial. Sporadic ALS, accounting for 90% to 95% of cases, occurs randomly without any known cause or family history. Familial ALS, affecting a smaller number of individuals, is believed to be inherited. Due to the majority of ALS cases being sporadic in nature, lacking an evident genetic predisposition, identifying a singular cause or mechanism underlying the disease has proven to be a complex and challenging task. One prevalent theory suggests that ALS results from a combination of genetic susceptibility and environmental factors. Dysfunction in cellular processes, particularly protein misfolding involving the protein TDP-43, and inflammation are thought to play pivotal roles in the disease's progression. However, the exact interplay of these factors remains incompletely understood. ALS is notorious for its delayed diagnosis, with individuals typically receiving this life-altering news approximately 12 months after the onset of their first symptoms. Following diagnosis, patients typically face a survival timeline ranging from 3 to 5 years. This delay in diagnosis can be attributed to a multitude of factors.
Firstly, ALS is relatively rare, with only limited cases around the world. Moreover, the presentation of symptoms and the progression of the disease vary widely among patients. Early signs and symptoms of ALS are often subtle and can be mistaken for signs of aging or other neuromuscular conditions. Additionally, there is no single definitive test for ALS, making it challenging to arrive at a rapid diagnosis. While genetic testing can provide answers for some cases, for many others, clinicians must rely on a comprehensive evaluation of clinical history, electromyography, neuroimaging, and physical examinations.
A. The Impact of Delayed Diagnosis
Delayed diagnosis of ALS has significant implications for patients. It postpones their access to multidisciplinary care, which has been shown to enhance their quality of life and extend survival. Furthermore, it hinders the initiation of disease-modifying treatments. Historically, the only FDA-approved treatment for ALS was riluzole, which provided only a modest extension of life expectancy. However, recent developments in therapeutic options, including the approval of edaravone, have heightened the importance of early diagnosis.
In 2017, the FDA approved edaravone, an antioxidant drug administered intravenously (IV), as a treatment for ALS. Clinical trial participants receiving edaravone experienced a significant reduction in the decline of daily function, indicating its potential to slow disease progression. The FDA's recent approval of an oral formulation of edaravone in 2022 offers the promise of more accessible at-home treatment, removing the need for IV administration.
Beyond the immediate benefits of approved treatments, early diagnosis is crucial for enrolment in clinical trials. Clinical trials often require a definitive or probable ALS diagnosis for eligibility, and there are currently over 100 products in development for ALS management, including several investigational therapies in phase 3 trials. Delayed diagnosis not only deprives patients of potential access to these innovative treatments but also hinders the research and development process for new, potentially effective therapies.
Recent advancements in AI and ML technologies have revolutionized ALS research, offering new hope for improved diagnosis, treatment, and ultimately, a cure. These technologies have significantly enhanced our ability to tackle the complex facets of the disease:
- Genomic Analysis: AI algorithms can analyse vast genetic datasets, identifying genetic markers associated with ALS susceptibility and progression. This enables a deeper understanding of the genetic underpinnings of both familial and sporadic ALS cases.
- Drug Discovery: ML models can screen vast chemical libraries to identify potential drug candidates that target specific pathways involved in ALS. This accelerates the drug development process and offers a more personalized approach to treatment.
- Patient Stratification: AI-driven data analysis can segment ALS patients into subgroups based on their unique genetic and clinical profiles. This aids in tailoring treatment approaches, optimizing patient care, and improving clinical trial outcomes.
- Disease Modelling: AI-powered simulations and neural networks help recreate the complex cellular and molecular interactions involved in ALS. This facilitates the testing of various hypotheses and potential interventions.
B. Role of Genetics
Genetics plays a critical role in ALS. While most cases of ALS are not directly inherited and occur sporadically, a significant proportion of ALS cases have a genetic component. One of the most common genetic mutations associated with ALS is the expansion of the C9orf72 gene's hexanucleotide repeat. This mutation disrupts normal cellular processes and is responsible for a substantial proportion of both familial and sporadic ALS cases.
C9orf72 is a gene found in our DNA, and it plays a crucial role in maintaining the health of nerve cells. In some individuals with ALS, a mutation in the C9orf72 gene leads to a unique problem: the gene has an abnormally expanded hexanucleotide repeat, meaning a particular sequence of six DNA letters is repeated too many times.
This expanded repeat in C9orf72 is a key genetic factor in certain cases of ALS. It disrupts normal cellular processes, affecting the production of essential proteins. The accumulation of these abnormal proteins in nerve cells contributes to the damage and death of these cells, leading to the progressive muscle weakness and loss of motor function characteristic of ALS.
In fact, C9orf72 mutations are among the most commonly known genetic causes of ALS. Understanding the role of C9orf72 in ALS has been a significant breakthrough in our knowledge of this devastating disease. It has paved the way for research into targeted therapies that aim to address the specific problems caused by this genetic mutation, offering hope for potential treatments for ALS in the future.
In this article, we use the Machine learning techniques to analyse the genetic mutations of C9orf72 to identify the severity of the disease.
II. P BACKGROUND STUDY
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disease characterized by the progressive degeneration of motor neurons, leading to muscle weakness and, ultimately, respiratory failure. One of the significant challenges in managing ALS is the delay in diagnosis, primarily attributed to the absence of a definitive biomarker. This literature review explores recent research efforts aimed at enhancing the diagnosis, understanding the genetic factors contributing to ALS, and addressing the need for early detection.
Vidovic et al. (2023) conducted a comprehensive review of 170 articles to shed light on the diagnosis of ALS. They emphasized the urgency of early and precise diagnosis and discussed several diagnostic approaches, including genetic testing, fluid biomarkers, and advanced imaging techniques. This review underscores the potential of these methods to improve diagnostic accuracy and prognosis for individuals with ALS.
Van Daele et al. (2023) conducted a study focusing on the genetic variability in sporadic ALS (sALS). Analyzing whole genome sequencing data from 6013 sALS patients and 2411 matched controls, the study aimed to gain insights into the genetic factors contributing to sALS. The findings provide a comprehensive overview of genetic variation in a large sALS cohort and underscore the complexity of the genetic factors involved in the disease. This research has implications for genetic counseling and the development of gene panels for ALS diagnosis.
A recent study by Raghav et al. (2022) investigated the presence of gene fusion events associated with ALS. This research represents a novel exploration of gene fusion events in the context of ALS, an aspect that has not been extensively studied. The findings of this study suggest that gene fusion events, particularly those unique to ALS cases, may contribute to the genetic factors underlying ALS pathogenesis. Further research is required to elucidate the mechanisms and implications of these gene fusion events in the context of ALS (Raghav et al., 2022).
Mitsumoto et al. (2022) discussed the challenges associated with the diagnosis of ALS, a disease often diagnosed approximately 12 months after the onset of new progressive weakness. They highlighted the importance of recognizing that ALS begins long before symptoms appear and explored the potential benefits of early diagnosis, including earlier therapeutic interventions, cost savings, prolonged survival, and reduced psychological distress among patients.
Bram et al. (2019) reported on a study involving the development of a PCR assay to analyze the hexanucleotide repeat region in the C9orf72 gene, associated with both ALS and frontotemporal dementia. The research aimed to provide comprehensive genotyping of this region in 2095 ALS samples, with the potential to improve genetic testing accuracy and further our understanding of ALS and related disorders.
In the context of ALS clinical trials, Fournier (2022) discussed key considerations essential for designing effective trials. These considerations encompassed the selection of reliable outcome measures, statistical techniques to optimize trial efficiency, the development of biomarkers for target engagement, inclusion of diverse patient populations, and addressing challenges related to access to experimental treatments. These strategies collectively aim to enhance patient outcomes and expedite treatment validation.
DeJesus-Hernandez et al. (2011) made a groundbreaking discovery concerning the expanded GGGGCC hexanucleotide repeat in the C9ORF72 gene. This repeat expansion was found to be strongly associated with a rare form of frontotemporal dementia (FTD) and ALS, highlighting its significance as a major genetic factor in both diseases.
An et al. (2018) explored the possibility of using Convolutional Neural Networks (CNNs) to automatically detect ALS at an early stage based on highly intelligible speech signals. While their findings show promise, the authors acknowledged the need for further research and larger datasets to improve and validate this approach.
This literature review provides a comprehensive overview of recent research efforts aimed at enhancing ALS diagnosis, understanding its genetic underpinnings, and improving early detection methods. These advancements contribute to our ongoing efforts to address the challenges associated with ALS diagnosis and management.
III. METHODOLOGY
In this study, we have taken the effect of mutation of C90rf72 gene with parameters being its two alleles comprising the gene loci, and indels caused. Indels, short for "insertions" and "deletions," are types of genetic mutations or alterations that involve the insertion of one or more nucleotide bases into a DNA sequence or the deletion of one or more nucleotide bases from a DNA sequence.
The data set was obtained from The Coriell Institute for Medical Research containing the mutations count of Allele1 and Allele2 data (https://www.coriell.org/)
The objective of this study was for the machine learning algorithm to diagnose and distinguish between a normal healthy person, an intermediate stage where the person has a risk of ALS, and an affected individual at the stage of full mutation (FM).
The three target stages are described as follows:
- Full Mutation (FM): A "Full Mutation" typically refers to a C9orf72 hexanucleotide repeat expansion that has an abnormally large number of repeats. In the context of C9orf72-associated ALS, "FM" corresponds to a repeat length of several hundred to thousands of hexanucleotide repeats. (exponentially over 30)
- Level-2 Heading: An "Intermediate" repeat expansion generally refers to a C9orf72 hexanucleotide repeat length that falls between the normal range and the full mutation range (25-30 and above). The threshold for defining an "Intermediate" repeat length can vary, but it is typically less than the repeat length associated with "FM."
- Level-2 Heading: A "Normal" allele refers to the typical or non-expanded hexanucleotide repeat length in the C9orf72 gene. In the general population, most individuals have "Normal" repeat lengths with only a small number of hexanucleotide repeats (i.e., less than 30 repeats).
Some cases of ALS have been linked to genetic mutations, including insertions and deletions (indels) in certain genes. Mutations in the superoxide dismutase 1 (SOD1) gene are another well-known genetic cause of ALS. Some of these mutations can involve indels, where small insertions or deletions occur in the gene's DNA sequence. Indels and other types of genetic mutations can disrupt the normal function of genes and proteins involved in motor neuron function, leading to the neurodegeneration seen in ALS.
Research into the genetic basis of ALS, including the role of indels and other genetic variations, continues to advance our understanding of the disease and may eventually lead to targeted therapies or interventions.
We have incorporated the following machine learning algorithms to analyse the give data to provide and predict diagnosis based on the dataset.
A. Random Forrest
The Random Forest algorithm is a powerful and versatile machine learning technique that has gained popularity across various fields due to its ability to provide accurate predictions, handle large datasets, and mitigate overfitting.
- It is an ensemble learning method that combines multiple decision trees to make more accurate predictions. The fundamental idea behind ensemble learning is that aggregating the predictions of several models often yields better results than using a single model.
- Decision trees are the basic building blocks of a Random Forest. Each decision tree is trained on a random subset of the dataset (bootstrapped sample) and selects features at each node to split the data. The randomness in feature selection and data sampling helps prevent overfitting, a common issue with individual decision trees.
- Random Forest employs a technique called bagging (bootstrap aggregating), where multiple decision trees are trained on different subsets of the dataset. These subsets are created through random sampling with replacement, ensuring diversity among the trees. Bagging reduces the variance of the model and improves its generalization.
- At each node of a decision tree, a random subset of features is considered for splitting. This feature selection randomness further enhances the diversity of individual trees. It also helps in identifying the most important features for making accurate predictions.
B. Support Vector Machine
- SVM is known for its emphasis on finding the hyperplane that maximizes the margin between different classes. Maximizing this margin leads to better generalization and increased resistance to overfitting
- Decision trees are the basic building blocks of a Random Forest. Each decision tree is trained on a random subset of the SVM can efficiently handle nonlinear datasets by transforming them into a higher-dimensional space using kernel functions. Kernel functions allow SVM to capture complex relationships in the data without explicitly mapping it into higher dimensions.
- SVM's training process focuses on optimizing the position and orientation of the hyperplane with respect to the support vectors (i.e., data points closest to the hyperplane.)
C. k-NN (k Nearest Neighbour)
k-NN is considered an instance-based learning algorithm because it memorizes the entire training dataset instead of building a model.
- Predictions are made by finding the k-nearest data points to the test instance and aggregating their information. To make predictions, k-NN measures the distance (commonly using Euclidean distance) between the test data point and all data points in the training set.
- It selects the k-nearest neighbours based on the calculated distances. The choice of the hyperparameter k defines the number of nearest neighbours to consider when making predictions. A smaller k value (e.g., 1 or 3) makes predictions sensitive to local variations, while a larger k value (e.g., 5 or 10) provides more robustness but may overlook fine-grained patterns.
- In classification tasks, k-NN assigns the class label that is most common among the k-nearest neighbours. In regression tasks, it computes the average (or another aggregation) of the target values of the k-nearest neighbours as the prediction.
The following section describes the implementation results.
IV. RESULTS AND DISCUSSION
The proposed methodology was applied to the specified dataset using the Python environment, and the dataset was sourced from The Coriell Institute for Medical Research, which contains information on the mutation counts of Allele1 and Allele2. You can access the dataset at https://www.coriell.org/.
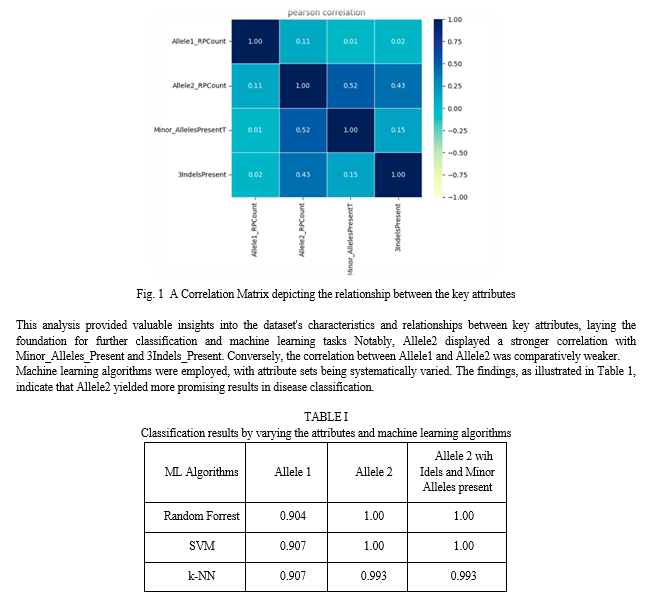
Upon conducting exploratory data analysis, it was determined that the dataset comprises 2095 samples with 11 attributes. Among these attributes, the most pertinent ones for classification were identified as Allele1 Count, Allele2 Count, Minor_Alleles Present, and 3IndelsPresent. These attributes were chosen due to their relevance in the classification task.
The target attribute in the dataset encompasses three classes, specifically Normal, Int, and FM, representing varying levels of disease severity. To gain insight into the relationships between these selected attributes, a correlation matrix was generated, and the results are illustrated in Fig. 1.
V. ACKNOWLEDGMENT
The author would like to extend their sincere gratitude to Dr. Hari Murthy, their esteemed biology professor, and Dr. Irene Getzi S, their mother, for their invaluable guidance, unwavering support, and expertise throughout the course of this research.
Conclusion
This study highlights the promising role of machine learning algorithms in diagnosing ALS based on C9orf72 gene mutations and indels. The results indicate a high level of accuracy in distinguishing between normal, intermediate, and full mutation stages of ALS. Early diagnosis is crucial for better patient outcomes and access to emerging treatments. These findings contribute to the ongoing efforts to combat ALS and underscore the potential of machine learning in advancing diagnostic precision and personalized treatment for neurodegenerative diseases like ALS. This research builds upon recent advancements in ALS diagnosis and genetics, providing valuable insights into the potential for machine learning to enhance early detection. Further research and validation are needed, but this study lays a foundation for future developments in ALS diagnosis and management.
References
[1] Vidovic, Maximilian, et al., Current State and Future Directions in the Diagnosis of Amyotrophic Lateral Sclerosis, Cells 12.5 (2023): 736. [2] Van Daele, Sien Hilde, et al., Genetic variability in sporadic amyotrophic lateral sclerosis, Brain (2023): awad120. [3] Raghav, Yogindra, et al., Identification of gene fusions associated with amyotrophic lateral sclerosis, medRxiv (2022): 2022-06. [4] Mitsumoto, Hiroshi, Edward J. Kasarskis, and Zachary Simmons., Hastening the diagnosis of amyotrophic lateral sclerosis, Neurology 99.2 (2022): 60-68. [5] Bram, Eran, et al., Comprehensive genotyping of the C9orf72 hexanucleotide repeat region in 2095 ALS samples from the NINDS collection using a two-mode, long-read PCR assay, Amyotrophic Lateral Sclerosis and Frontotemporal Degeneration 20.1-2 (2019): 107-114. [6] Fournier, Christina N., Considerations for Amyotrophic Lateral Sclerosis (ALS) Clinical Trial Design, Neurotherapeutics 19.4 (2022): 1180-1192. [7] DeJesus-Hernandez, Mariely, et al., Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS, Neuron 72.2 (2011): 245-256. [8] An, KwangHoon, et al., Automatic Early Detection of Amyotrophic Lateral Sclerosis from Intelligible Speech Using Convolutional Neural Networks, Interspeech. 2018.
Copyright
Copyright © 2023 Hans Elkan Sam C. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET56069
Publish Date : 2023-10-09
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

