Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- References
- Copyright
A Comparison on Supervised and Semi-Supervised Machine Learning Classifiers for Gestational Diabetes Prediction
Authors: Lokesh Kola, Vigneshwar Muriki
DOI Link: https://doi.org/10.22214/ijraset.2021.39434
Certificate: View Certificate
Abstract
Diabetes is the deadliest chronic diseases in the world. According to World Health Organization (WHO) around 422 million people are currently suffering from diabetes, particularly in low and middle-income countries. Also, the number of deaths due to diabetes is close to 1.6 million. Recent research has proven that the occurrence of diabetes is likely to be seen in people aged between 18 and this has risen from 4.7 to 8.5% from 1980 to 2014. Early diagnosis is necessary so that the disease does not go into advanced stages which is quite difficult to cure. Significant research has been performed in diabetes predictions. As time passes, challenges keep increasing to build a system to detect diabetes systematically. The hype for Machine Learning is increasing day to day to analyse medical data to diagnose a disease. Previous research has focused on just identifying the diabetes without specifying its type. In this paper, we have we have predicted gestational diabetes (Type-3) by comparing various supervised and semi-supervised machine learning algorithms on two datasets i.e., binned and non-binned datasets and compared the performance based on evaluation metrics.
Introduction
I. INTRODUCTION
The main cause of diabetes is due to high sugar levels in the blood. There is no permanent cure for diabetes. However, it can be prevented by early diagnosis. In recent years, the hype for Machine Learning is increasing in disease prediction especially during COVID-19 times. In the present scenario, it is difficult for patients to consult doctors. Research [1] shown that has shown that the occurrence of diabetes is more likely in adults aged 18 and above has risen from 4.7% to 8.5% from 1980 to 2014. By 2030, diabetes [2] is likely to be declared as the 7th major cause of death. Statistical results in 2017 shown that 451 million people are suffering from diabetes. If diabetes goes untreated, the count increases to 693 million by 2045 [3].
There are mainly three types of diabetes. Type-1 diabetes [4] is due to a lack of insulin. Type-2 diabetes [4] is due to the ineffective use of insulin. Type-3 diabetes also called gestational diabetes [5] is only seen in pregnant women without any previous history of diabetes. It is generally seen in women between the 24-28th week of pregnancy. If diabetes goes untreated it leads to blindness, heart stroke, nerve damage, chronic kidney diseases, etc.
In the contemporary era, Machine Learning (ML) and Artificial Intelligence (AI) has been increasingly used in healthcare. The computers are provided with the unstructured data and patterns are detected from it and thus predictions are made using the data. Diagnosing diabetes is a herculean task considering the complications. To make predictions accurate, effective analysis needs to be performed on medical data. ML and AI has the capability to detect such diseases. Depending entirely on technology is not the right way to diagnose a disease. Medical expertise should be involved in the diagnosis of diabetes as there are a lot of factors to be taken into consideration.
Most of the research [6] [7] [8] in diabetes used algorithms like Decision Tree (DT), Random Forest (RF), Support Vector Machine (SVM), Bernoulli Naive Bayes (BNB), Gaussian Naive Bayes (GNB), and Laplacian Support Vector Machine (LapSVM). LapSVM algorithm for diabetes prediction has produced good accurate results. Therefore, those algorithms were selected. Laplacian Support Vector Machine has been performed as semi-supervised learning while the rest are supervised.
In this paper PIMA Indians Diabetes Dataset was used to predict gestational diabetes. The dataset consists of women aged 21 years and above and is suffering from Type-3 diabetes. The dataset has been pre-processed and Feature selection was performed on the dataset to select the critical features impacting gestational diabetes. A new dataset has been created from the existing one by binning some of the important features. The two datasets (non-binned and binned) were used separately for training and testing the Machine Learning algorithms. Additionally, hyperparameter tuning was performed. The comparison between algorithms was performed based on accuracy, precision, recall, and f1-score.
II. METHOD
A. Data Collection
The dataset used in this paper was PIMA Indians Diabetes Dataset (PIDD). It consists of female patients aged 21 years and above from PIMA Indians Heritage. The dataset comprises 768 instances of which 500 patients are non-diabetic (represented by outcome 0) while the rest of them are diabetic (represented by outcome 1) which shows the imbalanced nature. There are eight features in the dataset. They are described as follows:
- Pregnancies: Denotes the number of times patient is pregnant
- Glucose: Plasma glucose concentration 2 hours in an oral glucose tolerance
- Blood Pressure: Diastolic blood pressure (mm Hg)
- Skin Thickness: Triceps skinfold thickness (mm)
- Insulin: 2-Hour serum insulin (mu U/ml)
- Body Mass Index (BMI): Body Mass Index = (weight in kg/ (height in m)2)
- Diabetes Pedigree Function: Diabetes history in relatives
- Age: Age in years
The PIDD dataset is obtained from Kaggle open-source database [9]. The second dataset created from the existing one by categorizing Age and BMI attributes. Data preprocessing was performed on the dataset to find missing values, null values, duplicated rows. Further, outliers were detected in each feature using box plot. Each attribute comprises of outliers. The quantile method was used to set the minimum and maximum threshold for Pregnancies. Since it is the only outlier found in the box plot because the value the wrongly entered in the dataset. While the rest of them are valid outliers as they aid in detecting the gestational diabetes. More the amount of data present, the better predictions can be made by the model.
B. Feature Selection
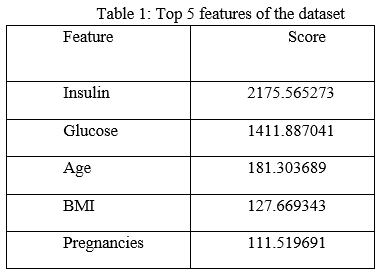
The features which show an impact on the target variable are selected. The SelectKBest method uses the chi-square test for selecting optimal features. The attributes with higher chi-square scores were considered to be the features showing the maximum impact on the target variable. The top five features were taken as optimal features based on the scores which are provided in decreasing order and then used for training the models. The scores for top five features were shown in Table 1.
The method which was discussed until now is the same for both datasets. Now the experiment has been performed in two parts. One experiment by performing binning on the dataset and the other without binning.
C. Binning
From the original dataset, the Age and BMI features were categorized. The dataset consists of women aged 21 years and above. Therefore, the Age [10] was divided into three categories namely Young Adult (21 < Age < 39), Middle-aged Adult (40 < Age < 59) and Old aged (Age > 60). BMI [11] was divided into four categories namely Underweight (0 < BMI < 18.5), Normal (18.6 < BMI < 25), Overweight (25 < BMI < 30) and Obese (BMI >= 30).
D. One-Hot Encoding
One-hot encoding was used to represent categorical data expressively. Mostly ML algorithms cannot work directly with categorical data. The categories must be changed to numbers or binary vectors. This applies to both input and output variables.
Since there were three categories in age, therefore three additional columns will be added to the data frame. Each column describes each category. Each category was associated with a binary value (0 or 1). The same applies to BMI which has four categories. Therefore, four additional columns were added to the data frame. Each row represents a binary vector. This reduces the burden in classifying multi class attributes.
E. Train-Test Split And Handling Imbalance Data
In the dataset, the outcome label was the dependent variable (denoted by y) and the rest of them were independent (denoted by x). The training data comprises 80% and testing data of 20%. The number of non-diabetic patients were 500 (indicated by outcome 0) and 268 patients were diabetic (indicated by outcome 1) which shows the imbalance nature of the dataset. Therefore, Oversampling technique was used to mitigate the same. Oversampling [12] [13] was performed on both the binned and non-binned training datasets separately. The number of samples in both training sets were equally distributed. The data has increased with an equal number of the target value. Now the model cannot be completely biased. After performing oversampling, the number of samples in the training data increased.
F. Training And Testing The Models
The training and testing of the model had been performed separately for each dataset. The DT, FC, SVM, BNB, and GNB algorithms were imported initially from the sklearn library. Metrics such as accuracy, precision, recall, and f1-score were calculated for each model on both datasets.
The training and testing for the LapSVM model were different from all the other models. The dataset was split into 80% training data and 20% testing data. The LapSVM code in Python was found in GitHub [14]. The class was instantiated. All the methods in the LapSVM class can be accessed by its object. The training dataset was further divided into two equal parts. Now, one part was considered as labelled dataset and the other was unlabeled.
The target attribute was dropped from one trained dataset, which refers unlabelled data. Therefore, the class has the fit method, where the model will be trained by using a labelled dataset. In the fitting process, the LapSVM class has done the computing adjacent matrix, computing Laplacian graph, computing kernel metrics then the metrics was inverted and computing the alpha, beta finally. Then the test dataset was given for the method accuracy in the LapSVM class. The accuracy method will predict the values and measure the accuracy then returns the score. Similarly, precision, recall, and f1-score were calculated for the prediction.
G. Hyperparameter Tuning
The hyperparameters for every classifier were tuned by using the GridSearchCV (GSCV) method. Initially, the GSCV was imported and the required parameters for the GSCV were estimator, parameter_grid, scoring, cross-validation, n_jobs. The estimator is the ML classifier and parameter_grid are the parameters of the classifier. All available parameters were given to the GSCV, and the training dataset will be used to fit the GSCV to give the correct parameters, which is suitable for efficient prediction. Then best_score will return the accuracy on the training dataset.
Each model’s multiple parameters and stratified-10-fold cross validation are passed to the GSCV and then the training data was given to the GSCV. GSCV performs internal cross validation on 80% of the train data to set the hyperparameters. The GSCV returnes the accuracy score on train data. Then best_params_ command was used to get the best parameters for the models. Since LapSVM model was a bit different from the other models, manual hyperparameter tuning was performed to obtain the best parameters and in turn a high performance.
III. RESULTS
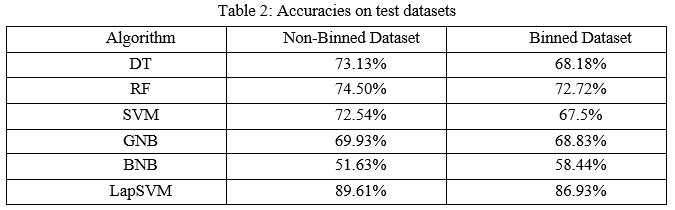
The models were trained and tested 10 times to obtain the average accuracies for both datasets. The accuracies were summarized in the below Table 2. The default parameters for some algorithms lead to better results. For the non-binned dataset, SVM and BNB obtained better accuracy with default hyperparameters. However, DT, RF, GNB, and LapSVM attained a higher accuracy with hyperparameter tuning. Likewise for binned dataset DT, RF, SVM scored higher accuracy with the default parameters. However, for LapSVM and GNB obtained higher accuracy after hyperparameter tuning. BNB scored the same accuracy with the default parameters and in hyperparameter tuning.
Accuracy is considered when TPs and TNs are crucial. Accuracy measures only the predictions which are correct. But the disadvantage is, it is not considered when the classes are imbalanced. Thus, to overcome such problems precision, recall, and f1-score are used to handle when classes are misclassified and imbalanced.
When FPs and FNs are important, f1-score is used. It is the harmonic mean of precision and recall which shows the misclassification of classes. Using harmonic mean, extreme values will be penalized. It measures trade-off between precision and recall.
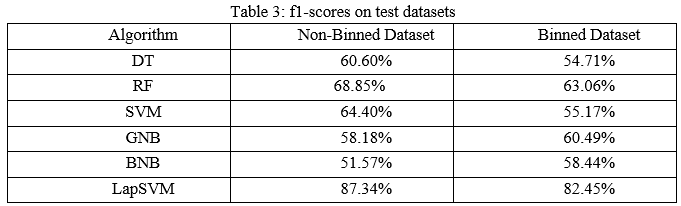
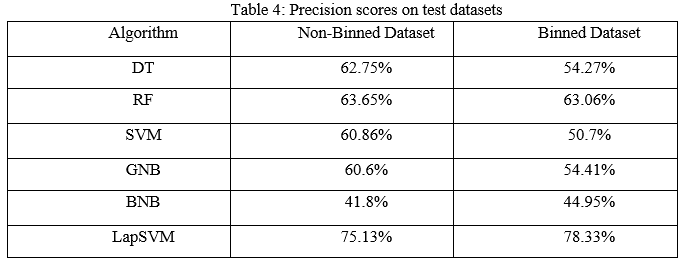
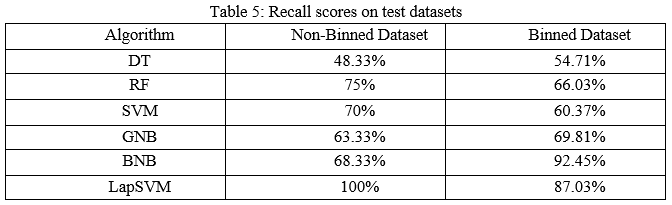
The precision scores of all the algorithms were described in Table 4. The LapSVM algorithms were scored high precision among other algorithms. For the non-binned dataset, the precision was 75.13% and 78.33% for the binned dataset. Higher precision means that more relevant results than irrelevant results are provided by an algorithm. Likewise, recall was also calculated for the algorithms and shown in Table 5. For the non-binned dataset, the recall score of LapSVM was 100%, which means LapSVM has no FNs. For the binned dataset, the high recall score was 92.45% for the BNB algorithm and LapSVM obtained 87.03%, which was the second highest recall score. When an algorithm has a high recall, it returns most of the relevant results whether irrelevant ones are also returned. The f1-scores were measured for selected algorithms and shown in Table 3 above. The LapSVM scored a high f1-score on both binned and non-binned datasets. High f1-score indicates that the model detects a smaller number of FPs and FNs. Note that in tables 2-5 the best performed algorithms are shown in bold.
Finally, the LapSVM, which is a semi-supervised learning technique has obtained the highest performance among the DT, RF, SVM, BNB, and GNB on both binned and non-binned datasets. The LapSVM was predicted with 87.34% accuracy on the non-binned dataset and 82.45% accuracy on the binned dataset. The reason was that the unsupervised learning methods learns new patterns which were undiscoverable in supervised learning methods. RF algorithm was the second best among other ML algorithms on both datasets. Comparing performances between binned and non-binned datasets, non-binned dataset produced better results. Hence the non-binned dataset is suitable for Type-3 diabetes prediction.
IV. FUTURE WORKS
In the future, diabetes can be performed with a precise dataset with all types of diabetes and consisting of both genders. Also, performance can be further improved by using Deep Learning techniques like Multi-Layer Perceptron (MLP), Artificial Neural Networks (ANN). ML algorithms worked well on training data but failed to work the same way on the new data due to overfitting. This can be solved by using the drop-out method. Dropout is only used during the training of a model and is not used when evaluating the skill of the model.
References
[1] J. Shou, L. Zhou, S. Zhu, and X. Zhang, “Diabetes is an Independent Risk Factor for Stroke Recurrence in Stroke Patients: A Meta-analysis,” Journal of Stroke and Cerebrovascular Diseases, vol. 24, no. 9, pp. 1961–1968, Sep. 2015, doi: 10.1016/j.jstrokecerebrovasdis.2015.04.004. [2] C. D. Mathers and D. Loncar, “Projections of Global Mortality and Burden of Disease from 2002 to 2030,” PLOS Medicine, vol. 3, no. 11, p. e442, Nov. 2006, doi: 10.1371/journal.pmed.0030442. [3] “IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045 - ScienceDirect.” https://www.sciencedirect.com/science/article/pii/S0168822718302031 (accessed Dec. 14, 2021). [4] “Diabetes in Western Pacific.” https://www.who.int/westernpacific/health-topics/diabetes (accessed Dec. 14, 2021). [5] K. Imam, “Gestational Diabetes Mellitus,” in Diabetes: An Old Disease, a New Insight, S. I. Ahmad, Ed. New York, NY: Springer, 2013, pp. 24–34. doi: 10.1007/978-1-4614-5441-0_4. [6] “Diabetes mellitus and its treatment | International Journal of Diabetes and Metabolism. 2005; 13 (3): 111-134 | IMEMR.” https://pesquisa.bvsalud.org/portal/resource/pt/emr-171007 (accessed Dec. 14, 2021). [7] “Long-Term Complications of Diabetes Mellitus | NEJM.” https://www.nejm.org/doi/full/10.1056/NEJM199306103282306 (accessed Dec. 14, 2021). [8] “Prospects for Research in Diabetes Mellitus | Diabetes | JAMA | JAMA Network.” https://jamanetwork.com/journals/jama/article-abstract/193527 (accessed Dec. 14, 2021). [9] “Pima Indians Diabetes Database.” https://kaggle.com/uciml/pima-indians-diabetes-database (accessed Dec. 14, 2021). [10] “Table 1 . Age intervals and age groups,” ResearchGate. https://www.researchgate.net/figure/Age-intervals-and-age-groups_tbl1_228404297 (accessed Dec. 14, 2021). [11] CDC, “Defining Adult Overweight and Obesity,” Centers for Disease Control and Prevention, Jun. 07, 2021. https://www.cdc.gov/obesity/adult/defining.html (accessed Dec. 14, 2021). [12] A. Liu, J. Ghosh, and C. Martin, “Generative Oversampling for Mining Imbalanced Datasets,” p. 7. [13] B. W. Yap, K. A. Rani, H. A. A. Rahman, S. Fong, Z. Khairudin, and N. N. Abdullah, “An Application of Oversampling, Undersampling, Bagging and Boosting in Handling Imbalanced Datasets,” in Proceedings of the First International Conference on Advanced Data and Information Engineering (DaEng-2013), Singapore, 2014, pp. 13–22. doi: 10.1007/978-981-4585-18-7_2. [14] H. Perrin, semi-supervised-learning. 2021. Accessed: Dec. 14, 2021. [Online]. Available: https://github.com/HugoooPerrin/semi-supervised-learning
Copyright
Copyright © 2022 Lokesh Kola, Vigneshwar Muriki. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET39434
Publish Date : 2021-12-14
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

