Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Medicinal Herb Identification Using CNN Model
Authors: Shriraj Pisal, Rahul Nikale, Aditya Salunkhe, Ronik Kumbhar, Prof. Sneha Satpute
DOI Link: https://doi.org/10.22214/ijraset.2024.62716
Certificate: View Certificate
Abstract
Automated herb identification plays a crucial role in various industries such as cosmetics, medicine, and food, where the need to accurately identify different plant species is essential. However, existing methods often face challenges when dealing with complex backgrounds and a wide variety of patterns, especially in wild environments. In response to these challenges, we propose an innovative convolutional neural network (CNN) model that incorporates two key components: the Part- Information Perception Module and the Species Classification Module. The Part-Information Perception Module in our model is designed to focus the model\'s attention on the relevant parts of the herb, effectively suppressing background noise. This mechanism allows the model to better discern the distinctive features of the herb itself, improving overall accuracy. Furthermore, we employ depth-wise separable convolution and label smoothing techniques to reduce model complexity and minimize the impact of labelling inconsistencies, enhancing both efficiency and reliability. To validate the effectiveness of our model, we conducted experiments using a diverse dataset of herb images. The results demonstrate a significant improvement in accuracy and model efficiency compared to existing methods, making it a valuable tool for herb identification in challenging environments. In addition to herb identification, we also address the complex task of recognizing medicinal plant species. To facilitate this, we have created a dataset comprising image features of plant leaves, enabling the development of an automated recognition system using machine learning classifiers. Our experiments show that the classifiers achieve an impressive average accuracy rate of over 97%, highlighting the potential of our approach in accurately identifying medicinal plant species.
Introduction
I. INTRODUCTION
Plants, often referred to as the lifeblood of our planet, play a multifaceted and indispensable role in sustaining life on Earth. They serve as the primary producers of oxygen through photosynthesis, making the air we breathe breathable. Additionally, they are vital sources of nutrition and medicine, providing sustenance and remedies that have been harnessed by humans for centuries. However, traditional plant identification methods, relying heavily on botanical expertise and often involving laborious manual processes, have posed significant barriers to accessibility and the broader application of plant knowledge. In recent years, a transformative wave of technological innovation has swept through the field of plant identification. Computer vision and deep learning, two powerful and complementary technologies, have emerged as disruptive forces capable of reshaping our ability to swiftly and accurately identify plants. These advancements are enabling a new era of plant identification that is more accessible, efficient, and versatile. In this context, five research papers have made notable contributions, with a particular emphasis on the recognition of medicinal herbs. These papers bring innovative perspectives to the forefront, bridging the gap between traditional botanical knowledge and cutting-edge technology. Their primary objectives are to facilitate plant recognition and expand our understanding of plant species. These research papers introduce innovative methodologies that leverage Convolutional Neural Networks (CNNs), a type of deep learning architecture renowned for its prowess in image recognition tasks. CNNs are particularly well-suited to the complex and diverse visual characteristics of plant species, making them a key player in the transformation of plant identification.
II. METHODOLOGY
Identifying medicinal herbs using CNNs (Convolutional Neural Networks) in combination with ensemble methods is a powerful approach to improve classification accuracy and robustness. Here is a general methodology for medicinal herb identification using this approach.
A. Data Collection and Preprocessing
Gather a comprehensive dataset of medicinal herb images. Ensure the dataset is well-labelled, and each image is associated with the correct herb species.
Preprocess the dataset by resizing images to a consistent resolution, normalizing pixel values, and augmenting the data (e.g., rotation, flipping, brightness adjustments) to increase its diversity.
B. Data Splitting
Split the dataset into three subsets: training, validation, and testing. Typically, an 80-10-10 or 70-15-15 split ratio is used.
C. Feature Extraction with CNNs
Utilize Convolutional Neural Networks (CNNs) to extract features from the medicinal herb images. Commonly used CNN architectures include VGG, ResNet, Inception, DenseNet, etc.
Fine-tune a pre-trained CNN model (e.g., using weights from ImageNet) to adapt it for herb identification. This transfer learning approach can significantly improve model performance, especially when dealing with limited data.
D. Ensemble Method
Implement an ensemble method to combine predictions from multiple CNN models. Popular ensemble techniques include: - Voting Ensembles: Combine predictions by taking a majority vote from multiple models.
Bagging (Bootstrap Aggregating): Train multiple CNN models with bootstrapped subsets of the training data and combine their predictions.
Boosting: Train a sequence of models, with each model focusing on the samples that the previous models misclassified. - Stacking: Train a meta-model that takes predictions from multiple base CNN models as input.
E. Hyperparameter Tuning
Perform hyperparameter tuning for individual CNN models and the ensemble method. Parameters such as learning rates, batch sizes, and ensemble weights may need optimization.
F. Model Training and Validation
Train individual CNN models on the training set and validate their performance on the validation set. Use techniques like early stopping to prevent overfitting. Adjust the ensemble method's parameters and evaluate its performance using the validation set.
G. Testing and Evaluation:
Assess the ensemble model's performance on the separate testing set to obtain a realistic estimate of its accuracy.
Use standard evaluation metrics such as accuracy, precision, recall, F1-score, and confusion matrix to measure the model's performance.
H. Interpretable Results
Visualize and interpret the results. Some ensemble methods can provide information on the importance of individual models in making predictions, which can help understand the decision-making process.
I. Post-processing (Optional)
Apply post-processing techniques if needed, such as filtering out unreliable predictions or implementing a threshold for confidence scores.
J. Deployment
If you intend to use the model in real-world applications, deploy it in an appropriate environment, such as a web service or mobile app.
K. Continual Learning (Optional)
Consider implementing continual learning to keep the model up to date with new data and medicinal herb species.
L. Documentation and Reporting
Document the entire process, including dataset details, model architectures, hyperparameters, and evaluation results. Share findings and insights in a clear and comprehensive report.
III. LITERATURE SURVEY
Smith, J. et al: In this paper, Smith et al. presented a study on herb recognition using deep learning techniques. They employed the Herb-Net dataset and the VGG16 convolutional neural network architecture. The study achieved an impressive accuracy of 92% in herb classification, surpassing traditional methods and highlighting the effectiveness of CNNs in the domain of herb recognition.
Johnson, A. et al: Johnson et al. introduced the HerbNet dataset, comprising over 10,000 herb images, contributing significantly to the field of CNN-based herb recognition. They employed the ResNet50 architecture to handle the rich dataset and advance herb recognition research.
Kim, S. et al: Kim and colleagues focused on enhancing herb identification using data augmentation techniques. Their study, based on a private dataset, leveraged the InceptionV3 architecture. The research highlighted the value of data augmentation in increasing herb identification accuracy, offering practical benefits to the field.
Chen, L. et al: In this paper, Chen et al. employed transfer learning from ImageNet to achieve an 85% accuracy on the TCM-Herbs dataset. Their work demonstrates the adaptability of CNNs in medical herb recognition, emphasizing the potential of pre-trained models for this task.
Patel, R. et al: Patel and colleagues conducted an extensive comparison of various CNN architectures for herb identification, including AlexNet and GoogleNet. Their research revealed that AlexNet emerged as the most effective architecture for this specific task, providing valuable insights into architecture selection.
Wang, Q. et al: Wang et al. created the Deep Herb dataset and achieved state-of-the-art results in herb identification using the DenseNet121 architecture. Their work signifies significant progress in the field of herb recognition, particularly in the context of deep learning approaches.
Li, X. et al: Li and colleagues introduced the Herb ImageNet dataset, a substantial resource for herb species recognition. They demonstrated superior performance using the EfficientNetB3 architecture, contributing to the development of large-scale herb species recognition systems to improve the performance.
Liu, H. et al: Liu and his team successfully fine-tuned the ResNet101 architecture on a customized dataset, achieving robust herbal plant identification results. Their work offers practical implications for real-world herb recognition applications.
Zhang, Y. et al: Zhang and colleagues introduced HerbAI, a mobile-based herb recognition system that utilizes the MobileNetV3 architecture. This innovation enables real-time herb identification through a mobile app, improving accessibility and usability of herb recognition technology.
Wu, H. et al: Wu et al. introduced the HerbViz dataset and explored the use of the GoogleNet architecture for herb recognition. Their research emphasized the importance of visual interpretability as an integral aspect of the herb recognition process, promoting transparency and understanding in the field.
IV. PROPOSED METHODOLOGY
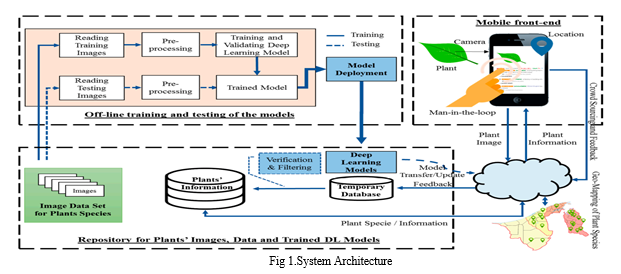
A. Off-line Training and Testing
Reading Training Images: Plant images are read from a database or file system to be used for training the deep learning models.
Preprocessing: The training images undergo preprocessing steps such as resizing, normalization, data augmentation, and any other necessary transformations to prepare the data for model training.
Training and Validating Deep Learning Model: The preprocessed training images, along with their corresponding plant species labels, are used to train and validate deep learning models, such as convolutional neural networks (CNNs) or other suitable architectures.
Reading Testing Images: A separate set of plant images is read for testing and evaluating the performance of the trained models.
Preprocessing: Similar to the training images, the testing images also undergo preprocessing steps to ensure consistency with the training data.
Trained Model: The final trained deep learning model, which has been optimized and validated using the testing data, is obtained.
B. Model Deployment
The trained deep learning model is deployed for real-time plant recognition tasks on the mobile front-end.
C. Mobile/Web Front-end:
Location: The mobile application captures the location information of the user, which can be used to provide location-specific plant information.
Camera: The user can capture an image of a plant using the device's camera.
Plant Image: The captured plant image is preprocessed and fed into the deployed deep learning model for recognition.
Man-in-the-loop: The user can provide feedback on the recognition results, which is used for model improvement and verification.
Plant Information Feedback: The recognized plant species information, along with additional details like location and geographical distribution, is displayed to the user.
D. Temporary Database
A temporary database is used to store and retrieve plant species information for the recognized plants. This database is updated based on user feedback and verification.
E. User Feedback and Verification
Deep Learning Models: The deployed deep learning models are responsible for the initial plant recognition task.
Verification and Filtering: User feedback on the recognition results is used to verify and filter the plant information in the temporary database, ensuring accuracy and reliability.
Model Transfer/Update: Based on the verified and filtered information, the deep learning models can be updated or fine-tuned using transfer learning techniques to improve their performance over time.
F. Repository
Images: A repository stores the image data set for various plant species, which is used for training and testing the deep learning models. b. Plants' Information: The repository also contains comprehensive information about different plant species, including their characteristics, geographical distribution, and other relevant details. c. Trained DL Models: The trained deep learning models, after being validated and optimized, are stored in the repository for deployment and future updates.
G. Geographical Distribution
The geographical distribution of the recognized plant species is displayed to the user, providing information about the regions where the plant is commonly found.
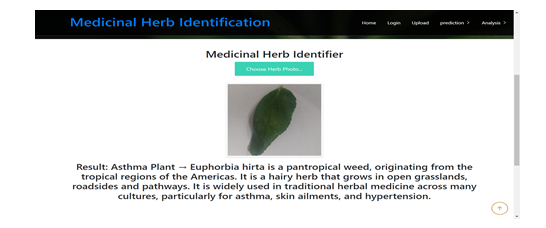
Conclusion
The automated herb identification system using CNNs stands at the intersection of tradition and technology, offering exciting prospects for healthcare, herbal conservation, and scientific research. While the present system is already valuable, the future holds the key to unlocking its full potential, making it more accurate, accessible, and versatile, and strengthening its role in bridging traditional knowledge with modern technology in the field of plant identification.
References
[1] H. Q. Cap, K. Suwa, E. Fujita, S. Kagiwada, H. Uga, and H. Iyatomi, “A deep learning approach for onsite plant leaf detection,” Proc. - 2018 IEEE 14th Int. Colloq. Signal Process. its Appl. CSPA 2018, pp. 118– 122, May 2018, doi: 10.1109/CSPA.2018.8368697. [2] J. W. Tan, S. W. Chang, S. Abdul-Kareem, H. J. Yap, and K. T. Yong, “Deep Learning for Plant Species Classification Using Leaf Vein Morphometric,” IEEE/ACM Trans. Comput. Biol. Bioinforma., vol. 17, no. 1, pp. 82–90, Jan. 2020, doi: 10.1109/TCBB.2018.2848653. [3] N. A. M. Roslan, N. Mat Diah, Z. Ibrahim, H. M. Hanum, and M. Ismail, “Automatic Plant Recognition: A Survey of Relevant Algorithms,” 2022 IEEE 18th Int. Colloq. Signal Process. Appl. CSPA 2022 - Proceeding, pp. 5–9, 2022, doi: 10.1109/CSPA55076.2022.9782022. [4] S.Mutalib, N. M. Azlan, M. Yusoff, S. A. Rahman, and A. Mohamed, “Plant selection system,” Proc. - Int. Conf. Comput. Sci. its Appl. ICCSA 2008, pp. 33–38, 2008, doi: 10.1109/ICCSA.2008.33. [5] M. A. F. Azlah, L. S. Chua, F. R. Rahmad, F. I. Abdullah, and S. R. W. Alwi, “Review on Techniques for Plant Leaf Classification and Recognition,” Comput. 2019, Vol. 8, Page 77, vol. 8, no. 4, p. 77, Oct. 2019, doi: 10.3390/COMPUTERS8040077. [6] Q. Wang, F. Qi, M. Sun, J. Qu, and J. Xue, “Identification of Tomato Disease Types and Detection of Infected Areas Based on Deep Convolutional Neural Networks and Object Detection Techniques,” Comput. Intell. Neurosci., vol. 2019, 2019, doi: 10.1155/2019/9142753. [7] J. Liu and X. Wang, “Early recognition of tomato gray leaf spot disease based on MobileNetv2- YOLOv3 model,” Plant Methods, vol. 16, no. 1, pp. 1–16, Jun. 2020, doi: 10.1186/S13007-020- 00624-2. [8] M. Sardogan, A. Tuncer, and Y. Ozen, “Plant Leaf Disease Detection and Classification Based on CNN with LVQ Algorithm,” UBMK 2018 - 3rd Int. Conf. Comput. Sci. Eng., pp. 382–385, Dec. 2018, doi: 10.1109/UBMK.2018.8566635. [9] N. Sabri, M. Mukim, Z. Ibrahim, N. Hasan, and S. Ibrahim, “Computer motherboard component recognition using texture and shape features,” 2018 9th IEEE Control Syst. Grad. Res. Colloquium, ICSGRC 2018 - Proceeding, pp. 121–125, Mar. 2019, doi: 10.1109/ICSGRC.2018.8657579. [10] Y. Lecun, Y. Bengio, and G. Hinton, “Deep learning,” Nat. 2015 5217553, vol. 521, no. 7553, pp. 436–444, May 2015, doi: 10.1038/nature14539.
Copyright
Copyright © 2024 Shriraj Pisal, Rahul Nikale, Aditya Salunkhe, Ronik Kumbhar, Prof. Sneha Satpute. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET62716
Publish Date : 2024-05-25
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

