Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Next Generation Sequencing and Insilico Identification of Human Anaplastic Lymphoma Kinase in Human Cancer with Biopython
Authors: Uma Kumari, Purnima Acharya
DOI Link: https://doi.org/10.22214/ijraset.2024.59267
Certificate: View Certificate
Abstract
In this comprehensive molecular investigation, the focus centers on the anaplastic lymphoma kinase (ALK) within the context of lung cancer. Specifically, utilizing the sample with the PDB code 3AOX, the study employs a multifaceted approach combining advanced bioinformatics tools and structural biology methodologies.Commencing with RasMol, the analysis of the protein sample reveals a complex network of 196 hydrogen bonds within the ALK structure. This foundational information sets the stage for subsequent investigations. Visual representation is meticulously addressed, with RasMol utilized for color-coding alpha helices in magenta, beta helices in yellow, and the remaining residues in white. The presentation mode emphasizes helices in red, sheets in yellow, and loops in green, providing a vivid depiction of the structural elements within the ALK protein.Moving beyond visualization, PyMOL is engaged to represent the surface form, providing insights into the interactions between ALK and its environment. This is particularly crucial in understanding the structural dynamics of ALK in the context of lung cancer, where the protein sample serves as a representative model.Active site identification in Chain A is pursued using tools such as PyMol, offering a glimpse into potential functional regions crucial for ALK\'s role in lung cancer pathogenesis. The study extends to biophysical aspects, incorporating protein-ligand docking studies with Mektovi and Almita through CB DOCK. This approach sheds light on the intricate molecular interactions between ALK and specific ligands, providing valuable information for targeted therapies.Structural validation becomes paramount in ensuring the reliability of the 3AOX sample. The ERRAT structure validation server evaluates the overall quality factor, while Procheck on the SAVES server scrutinizes the Ramachandran plot, ensuring the conformational consistency of ALK\'s backbone dihedral angles.Biopython scripts play a pivotal role in extracting and analyzing data related to the alpha-beta class representation within the CATH database. This bioinformatics analysis adds another layer of understanding to ALK\'s structural classification, offering insights into its role in lung cancer.
Introduction
I. INTRODUCTION
Anaplastic lymphoma kinase, often abbreviated as ALK, is a gene that provides instructions for making a protein that plays a role in the development and function of the nervous system(Robert, 2013). When we talk about the human ALK gene in the context of diseases, it's often associated with certain types of cancer(Bengt & Ruth, 2013).Over the past decade, groundbreaking advancements in cancer treatment have been underscored by the discovery and successful development of targeted therapies, particularly tailored for genetically defined subsets of patients(Hannaneh et al., 2024). Agents like imatinib, trastuzumab, lapatinib, and erlotinib have demonstrated remarkable efficacy in addressing tumors harboring specific genetic abnormalities(Toshimitsu et al., 2018). However, a persistent challenge remains, as the majority of human cancers resist currently available molecularly targeted agents. For instance, in non-small cell lung cancer (NSCLC), where only 10% of white patients exhibit an activating EGFR mutation sensitive to erlotinib, the remaining 90% with wild-type EGFR experience minimal therapeutic benefits(Lorenza & Federico, 2015).This comprehensive research delves into an exciting and promising avenue in targeted therapy for NSCLC—specifically focusing on lung cancers featuring anaplastic lymphoma kinase (ALK) fusion oncogenes. Utilizing state-of-the-art techniques such as Next-Generation Sequencing (NGS), the research aims to unravel the molecular intricacies of ALK-positive cancers and explore innovative treatment strategies.
The study, conducted across a diverse panel of human tumor-derived cell lines, leverages an automated platform to assess sensitivity to various molecularly targeted inhibitors.
Notably, molecular docking techniques are employed to understand the interaction between the inhibitors and the ALK protein. The identification of a subset of cells, including those derived from anaplastic large-cell lymphomas, non-small-cell lung cancers, and neuroblastomas, responsive to ALK inhibition underscores the potential clinical relevance of such targeted therapies. The specificity of the responses is further elucidated through the correlation with specific ALK genomic rearrangements, such as chromosomal translocations and gene amplifications(Itziar et al., 2008).
Moving beyond cell lines, the study extends its exploration to ALK-positive anaplastic large-cell lymphoma (ALK+ ALCL)(Xin-Rui et al., 2022). Employing advanced molecular techniques, including NGS(Uma & Shruti, 2023), the study offers a comprehensive analysis of fusion partners and presents a real-time quantitative reverse-transcription polymerase chain reaction method for monitoring ALK gene expression(Julie et al., 2017). Molecular docking simulations also contribute to understanding the binding interactions between ALK inhibitors and the unique ALK fusion proteins found in ALCL(Rahul et al., 2022).Employing BioPython's SeqIO module facilitates the extraction and manipulation of ALK sequence data, allowing for a detailed analysis of its composition(Jason et al., 2002). Additionally, the Bio.PDB module within Biopython proves essential for parsing PDB files, enabling the extraction of structural insights ofprotein (Simon et al., 2022). This structural information can be visualized using Biopython, enhancing our comprehension of ALK's three-dimensional conformation(Jian Li et al., 2017). Leveraging Bio.Entrez facilitates the retrieval of additional information from databases such as NCBI, contributing to a broader understanding of ALK's variations and associations. BioPython’s Phylo module offers a valuable avenue for phylogenetic analysis, shedding light on the evolutionary relationships of ALK across species(Eric Talevich et al., 2012). Furthermore, the integration of Biopython with machine learning libraries enables classification tasks, predicting various aspects of ALK based on existing data. In structural bioinformatics, Biopython can be coupled with tools like Autodock for molecular docking studies, predicting potential binding interactions with ligands(Priya & Uma, 2024). Automated report generation and statistical analysis, supported by Biopython, streamlines the summarization and interpretation of diverse data, providing a comprehensive overview of ALK's role in lung cancer(Uma & Keshav, 2023). The challenges of noninvasive detection of ALK rearrangements are addressed through the incorporation of capture-based sequencing in blood samples. This approach, alongside biocomputational analyses using Biopython, enhances the sensitivity for identifying driver fusion genes and monitoring tumor dynamics, including the emergence of drug resistance mutations.In the context of cancer immunotherapy, the paper examines acquired resistance to PD-(L)1 blockade in NSCLC(Justyna B?ach et al., 2021). Leveraging molecular profiling techniques, including NGS, the study sheds light on the clinical and molecular features of acquired resistance, providing insights into the persistently inflamed tumor microenvironment. Biopython scripts are employed for bioinformatics analysis, offering a systematic approach to interpreting large-scale genomic data(Priya & Uma, 2024).
Finally, this research discusses the pivotal role of receptor tyrosine kinases (RTKs) in cancer pathogenesis, with a particular focus on the anaplastic lymphoma kinase (ALK) as a novel tumorigenic player. The expression of ALK-RTK, its fusion proteins, and potential therapeutic strategies for ALK-positive neoplasms are explored, emphasizing the integration of cutting-edge technologies, such as NGS, molecular docking, and Biopython, in advancing our understanding and treatment of cancer.
II. METHODOLOGY
In this comprehensive molecular study, various bioinformatics and structural biology methodologies were employed to dissect and analyze different facets of a protein structure. Initial hydrogen bond analysis was conducted using RasMol, revealing 196 hydrogen bonds within the molecular structure. For visual representation, RasMol commands were employed to color code the alpha helices in magenta, the beta helices in yellow, and the remaining residues in white. Additionally, a presentation mode was utilized to highlight helices in red, sheets in yellow, and loops in green.The structural investigation extended to PyMOL for surface form representation, with a particular focus on visualizing the interactions between the amino acid and the ligand. Active site identification in Chain A was carried out using tools like Dali or CASTp to pinpoint potential functional regions.Bioinformatics analyses included BLASTP for sequence similarity, providing insights into the protein's homologous relationships and potential functional characteristics. Further exploration involved protein-ligand docking studies, with Mektovi and Almita being utilized through CB DOCK, offering detailed insights into the interactions between the protein and specific ligands.To validate the structural integrity, the ERRAT structure validation server was employed, yielding an overall quality factor for the molecular model. Ramachandran plot analysis, facilitated by the Procheck tool on the SAVES server, scrutinized the backbone dihedral angles for conformational consistency.Biopython scripting played a crucial role in extracting and analyzing data related to the alpha-beta class representation within the CATH database, providing a deeper understanding of the protein's structural classification.
This integrated approach not only delved into the intricacies of molecular interactions but also harnessed computational tools to unravel the structural and functional nuances of the studied protein.

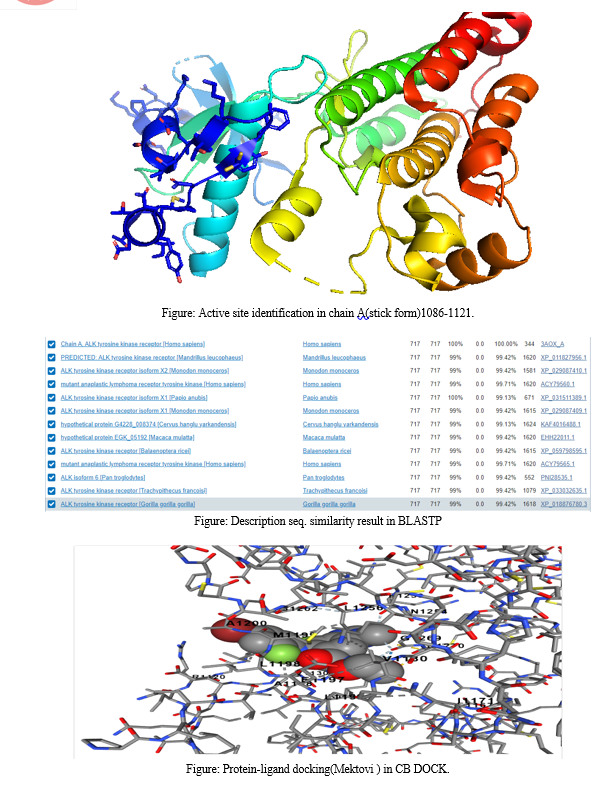
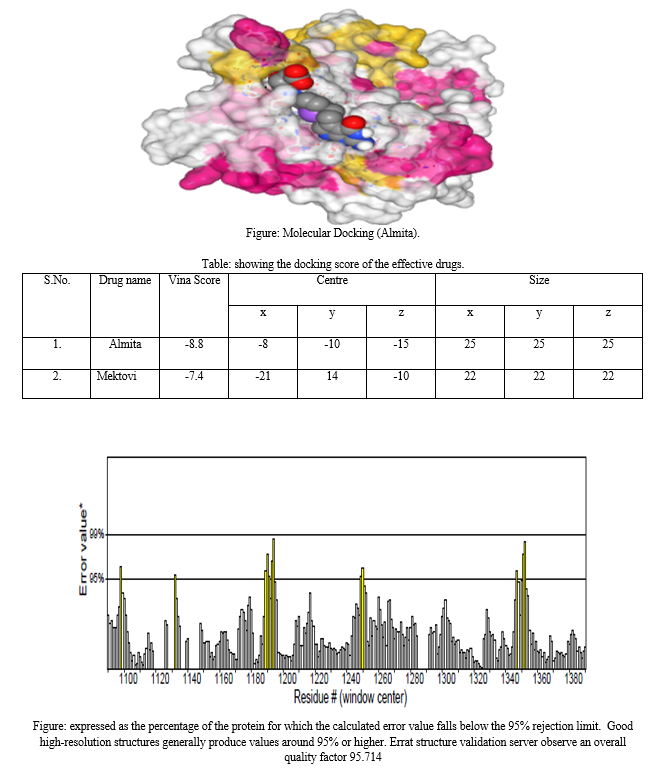


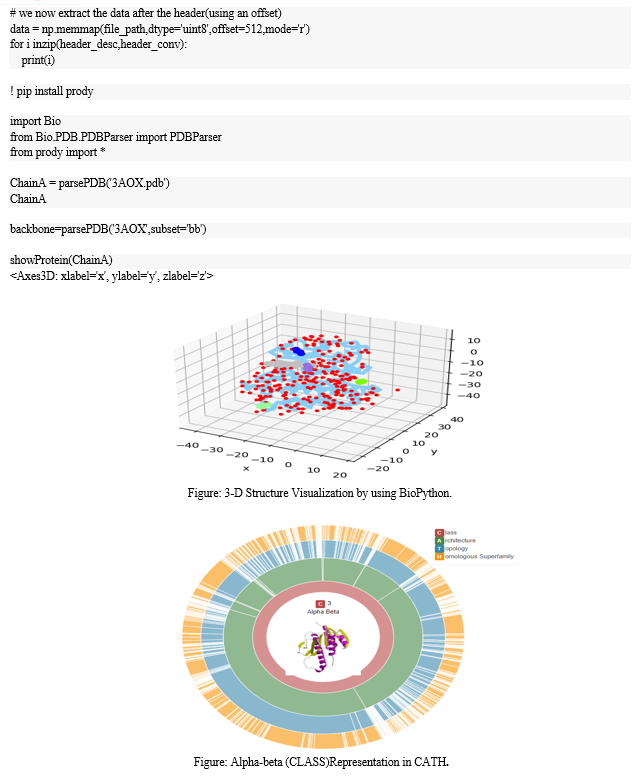
Conclusion
In conclusion, this research on the anaplastic lymphoma kinase (ALK) within the context of lung cancer, utilizing the 3AOX sample with the PDB code, provides valuable insights into the structural and functional aspects of ALK. The analysis of hydrogen bonds, coupled with visual representations and active site identification, enhances our understanding of ALK\'s molecular interactions. Protein-ligand docking studies offer potential therapeutic avenues, while structural validation ensures the reliability of our findings.Biophysical insights gained through protein-ligand docking studies with Mektovi and Almita enhance our understanding of ALK\'s interaction with specific ligands, offering potential avenues for targeted therapies. The structural validation process, including ERRAT and Procheck analyses, ensures the reliability and accuracy of the 3AOX sample, reinforcing the credibility of our findings. Moreover, the integration of Biopython scripts for alpha-beta class representation within the CATH database enriches our understanding of ALK\'s structural classification, providing context for its role in lung cancer. The application of these diverse methodologies converges to elucidate ALK\'s structural nuances, offering a holistic perspective on its involvement in lung cancer pathogenesis. The integration of Biopython scripts for structural classification adds a layer of context. Overall, this study contributes to the comprehensive understanding of ALK in lung cancer, laying the groundwork for further research and potential targeted interventions.
References
[1] Bengt Hallberg, R. H. (2013). Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. nature reviews cancer, 13, 685–700. [2] Eric Talevich, B. M. (2012). Bio.Phylo: A unified toolkit for processing, analyzing and visualizing phylogenetic trees in Biopython. BMC Bioinformatics, 13, 209. [3] Hannaneh Parvaresh. Ghazaal Roozitalab, F. G. (2024). Targeted Therapies in Non-Small Cell Lung Cancer: Comprehensive Insights and Future Directions. Biomedicines, 12(2), 297. [4] Itziar Salaverria, S. B.-G. (2008). Genomic profiling reveals different genetic aberrations in systemic ALK-positive and ALK-negative anaplastic large cell lymphomas. British Journal of Haematology, 140(5), 516-526. [5] Jason E. Stajich, D. B. (2002). The Bioperl Toolkit: Perl Modules for the Life Sciences. Genome research, 12(10), 1611-1618. [6] Jian Li, R. S. (2017). L1198F Mutation Resensitizes Crizotinib to ALK by Altering the Conformation of Inhibitor and ATP Binding Sites. IJMS, 18(3), 482. [7] Jr., R. R. (2013). Anaplastic lymphoma kinase (ALK): Structure, oncogenic activation, and pharmacological inhibition. Pharmacological Research, 68(1), 68-94. [8] Julie A. Vendrell, S. T.-L. (2017). Detection of known and novel ALK fusion transcripts in lung cancer patients using next-generation sequencing approaches. scientific reports, 7, 12510. [9] Justyna B?ach, K. W.-K. (2021). Failure of Immunotherapy—The Molecular and Immunological Origin of Immunotherapy Resistance in Lung Cancer. IJMS, 22(16), 9030. [10] KUMARI, P. A. (2024). Integrative Analysis Of Serine Dehydratase-Like (Sdsl) Gene In Liver Cancer Cells Through Ngs With Biopython. International Journal of Medicine and, 14(1), 11-22. [11] Lorenza Landi, F. C. (2015). Experience with erlotinib in the treatment of non-small cell lung cancer. Therapeutic Advances in Respiratory Disease, 9(4), 146-163. [12] Priya Arya, U. K. (2024). MOLECULAR DOCKING APPROACH FOR HUMAN ONCOLOGY RESEARCH WITH CHEMOINFORMATIC. Journal of Emerging Technologies and Innovative Research, 11(2), 658-666. [13] Rahul D. Jawarkar, P. S.-M.-H. (2022). QSAR, Molecular Docking, MD Simulation and MMGBSA Calculations Approaches to Recognize Concealed Pharmacophoric Features Requisite for the Optimization of ALK Tyrosine Kinase Inhibitors as Anticancer Leads. Molecules, 27(15), 4951. [14] Simon Jenni, J. A.-M. (2022). Visualizing molecular interactions that determine assembly of a bullet-shaped vesicular stomatitis virus particle. nature communications, 13, 4802. [15] Toshimitsu Yamaoka, S. K. (2018). Receptor Tyrosine Kinase-Targeted Cancer Therapy. IJMS, 19(11), 3491. [16] Uma Kumari, S. G. (2023). NGS and Sequence Analysis with Biopython for Prospective Brain Cancer Therapeutic Studies. International Journal for Research in Applied Science & Engineering Technology, 11(4), 3318-3329. [17] Uma Kumari. Keshav Saini, R. K. (2023). CADD APPROACHES FOR THE EARLY DIAGNOSIS OF LUNG CANCER. Journal of Clinical Otorhinolaryngology, Head, and Neck Surgery, 27(1), 5190-5200. [18] Xin-Rui Zhang, P.-N. C.-Y.-Y. (2022). Anaplastic Large Cell Lymphoma: Molecular Pathogenesis and Treatment. Cancers, 14(7), 1650.
Copyright
Copyright © 2024 Uma Kumari, Purnima Acharya. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET59267
Publish Date : 2024-03-21
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

