Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Plant Disease Detection Using YOLOv7 Algorithm
Authors: Rokkam Sai Praveen, Pothuri Sravani, Savalam Akhil, Tiyyagura Poojithaa Reddy, J M Babu
DOI Link: https://doi.org/10.22214/ijraset.2024.59272
Certificate: View Certificate
Abstract
Abstract: Plant diseases pose a serious danger to global food security and can result in huge financial losses for the agricultural sector. Earlier Plant disease detection and precise diagnosis are essential for putting management measures into place. Recent advancements in computer vision techniques have demonstrated encouraging outcomes in automating activities related to illness identification. This study uses the You Only Look Once (YOLOv7) object identification algorithm to present a novel method for plant disease diagnosis. The primary goal of this research is to create a reliable and effective system that can quickly and reliably identify plant diseases. YOLOv7, a highly accurate and speedy algorithm, will serve as the main underpinning for detection. The project\'s primary goal is to train the Yolov7 model to identify distinct citrus plant illnesses by using a large dataset that includes pictures of both healthy and diseased plants. This project categorizes leaf images recorded from a file or webcam into four categories: healthy, greening, blackspot, and canker. Early disease prediction allows farmers to take required security measures for their plants.
Introduction
I. INTRODUCTION
Agriculture is critical to the economy of every country. Agricultural advancements aim to meet increasing population demands. Agriculture needs modernization to thrive in today's environment. Crops are affected by both bacterial and fungal diseases. This significantly reduces farmers' production. Crop health is essential for achieving the highest yield. Detecting illnesses with the naked eye remains a complex task. Continuous monitoring of the farm is important. This is a tedious procedure. This can be costly, particularly for large farms. Agricultural professionals struggle to diagnose and address diseases due to their complexity. Farmers would benefit greatly from an automated approach for identifying plant illnesses. This technology can help farmers receive timely information and take required precautions. Plant diseases can injure several sections of the plant, including leaves, fruits and seeds. Plant diseases vary by plant body part. The leaves are the most vital portion of the plant. Plant disease may disturb the plant life cycle by affecting its leaves. Common leaf diseases include bacterial and fungal infections. Hence, early detection of plant disease is important.

Citrus crops are vulnerable to various diseases that can wreak havoc on their leaves, ultimately leading to significant losses in yield and quality. Diseases such as citrus greening, also known as Huanglongbing (HLB), and citrus canker primarily target the foliage of citrus trees. Citrus greening, caused by a bacterial pathogen spread by the Asian citrus psyllid, disrupts the vascular system of the trees, resulting in yellowing and mottling of leaves. Infected leaves often become misshapen and exhibit excessive dropping, severely impacting the tree's ability to photosynthesize and produce healthy fruit.
Citrus canker, on the other hand, manifests as raised lesions on leaves, which gradually enlarge and cause defoliation. As the disease progresses, affected trees become increasingly susceptible to environmental stresses, further exacerbating leaf loss and reducing overall productivity. Management of these leaf-targeting diseases often involves a combination of cultural practices, chemical treatments, and integrated pest management strategies to mitigate their spread and minimize losses in citrus crops. Early detection and prompt intervention are crucial to containing outbreaks and preserving the health of citrus orchards.
To effectively manage these disorders, it's crucial to have an automated method for recognizing and categorizing them. Several Deep Learning approaches have been developed to identify and classify plant diseases based on images. This paper utilizes YOLOv7, a deep learning approach for detecting citrus plant diseases. The paper is organized as follows: Section II considers influential work in a given field. Section III outlines the proposed methodology, technique, and phases for achieving desired goals. Section IV presents the results and interpretation of the proposed methodology. Section V presents the paper's conclusion.
II. REVIEW OF RELATED WORK
This section explains the variety of systems that use machine learning and deep learning techniques to identify plant leaf sickness. By using the CNN architecture known as LeNet, CNN is used to categorize tomato plant leaf photos based on the noticeable effects of the diseases [1]. The primary goal of the study is to find a way to identify tomato leaf disease as easily as possible with the least amount of computer resources while yielding results that are on track with state-of-the-art techniques. A method for identifying plant illnesses was put out by Ms. Deepa et al. .This method involved pre-processing the digital photographs of the plants and utilizing Support Vector Machines (SVM) to process them [2].
Keras Conv2D has been utilized by Faizan Akthar and colleagues for clustering [3]. Using an optical camera or similar devices, they have taken pictures of several leaf species and labeled them with the corresponding diseases in this method. The plant disease detection method was implemented by Zhaoyi Chen et al. [4] utilizing the YOLOv5. They employed the YOLOv5 model to identify the plant diseases accurately, with an accuracy of 86–87 %.
A method for achieving plant disease detection through the use of ConvNets to create a CNN was proposed by Marwan Adnan Jasim et al. [5]. Through processing the leaf, these earlier studies and research attempts studying plant disease detection were able to identify the diseases in the plant. YOLOv7 offers numerous benefits over alternative techniques for identifying plant diseases. First off, it performed better than other detectors in the CP dataset, as proved by its higher mAP score and significant margin of the result.
III. METHODOLOGY
The plant's leaves are frequently subject to various diseases. Humidity and other environmental factors are responsible for this. These diseases could be caused by viruses, fungi, or bacteria. These variations share similar characteristics, making them frequently challenging to identify. Farmers can avoid losses if they are detected early. The suggested method uses deep learning and the YOLOv7 algorithm to determine whether a plant is healthy or affected by diseases like Greening, Blackspot, and Canker.
The proposed technique for detecting plant diseases using the YOLOv7 algorithm involves multiple steps:
A. Dataset
Kaggle provided the dataset for this project, which includes photographs of both healthy and damaged leaves. Images with varying sizes may not be suitable with the YOLOv7 environment. The collection includes 587 photos of various sizes, all of which are saved in uncompressed png format and default to the RGB color space.
B. Pre-processing 1
To make the dataset’s photos compatible with YOLOv7 training and perform the extraction of results, they must be scaled to 640 * 640 resolution.
Data augmentation, which includes creating modified copies of the dataset using previous data, must be applied to the images in the dataset. It is essential because the suggested strategy depends on a lot of various kinds of data to generate accurate predictions. It resolves the overfitting issue and class imbalance.
Images in our dataset can be rotated and flipped to provide augmented data. As it adds more data, slightly altered versions of the original data are used. The number of photos in the dataset has increased to 817 after augmentation.
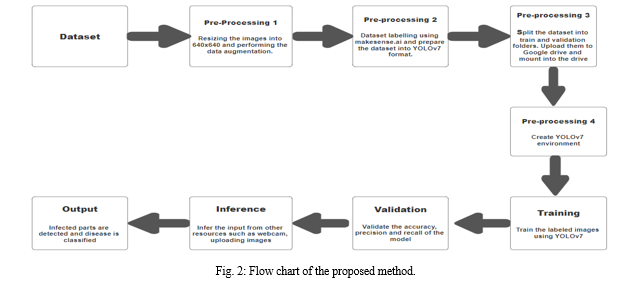
C. Pre-processing 2:
Developing supervised models with computer vision capabilities requires image labeling. Because it facilitates the training of deep learning models to recognize classes of objects within an image or to classify the entirety of the images. The image is labeled by recognizing and annotating different elements within it. For example, in a leaf, the disease can show in a variety of ways, including color, texture, and shape change as well as the presence of spots or blotches.
These must be taken into account, and the dataset’s 817 photos must be labeled. The makesense.ai software may be utilized to do this.
D. Pre-processing 3:
The dataset is divided into training and validation sets, which helps in determining how well the suggested model will apply to new, untested data. Additionally, it avoids overfitting, which occurs when a model produces good results on training data but not new examples. It facilitates evaluating the effectiveness of the learning approach.
The dataset is divided into our suggested model so that 80% of the photos are used for training and the remaining 20% are used for validation.
Training- 701 images
Validation- 116 images
To train the model, upload the dataset to the drive and access the Google Colabs virtual machine. To train the model and obtain the results, create a notebook. To access the data on our drive, we had to mount the drive to our notebook.
E. Pre-processing 4:
The next step is to install the dependencies needed to train the model in order to construct the YOLOv7 environment on our notebook.
When YOLOv7 is initialized with pretrained weights, it typically operates more efficiently. From the YOLOv7 repository, you can download them.
The YOLOv7 architecture, in general, is intended for real-time object recognition tasks and expands upon its predecessors. It is frequently used in conjunction with CSPDarknet53, which offers improved information flow through a Cross-Stage Partial (CSP) module. With YOLOv7’s quickness and adaptability, users can obtain precise results tailored to their individual requirements. One effective and adaptable option for real-time object identification applications is YOLOv7.
The general overview of the YOLOv7 architecture is:

- Input: The input provided to YOLOv7 is an image of size 640 x 640 and provided to train the model.
- Backbone network: A backbone network is usually used by YOLOv7 to extract feature maps from the input image. The image’s hierarchical information, from minute features to high-level semantics, is captured by the feature maps. For increased efficiency, it has CSPDarknet53 with a CSP module.
- Neck: The neck is in charge of integrating data from several backbone feature map levels. This integration increases the accuracy and helps the model capture context more accurately.
- Detection head or Prediction: Using the anchor boxes, it is in charge of predicting bounding boxes, class probabilities, and object confidences. The model can recognize objects of varying sizes since it is capable of predicting these values at different scales and resolutions.
- Output: YOLOv7 produces a set of bounding boxes as its final result, and each one has a class label and confidence score assigned to it.
F. Training
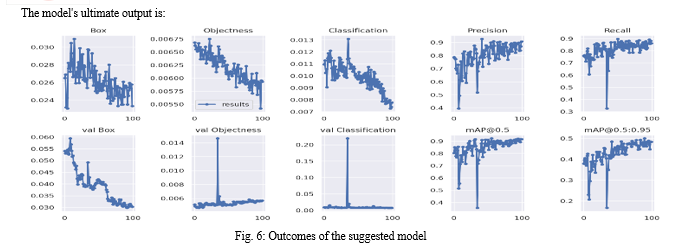
The model is trained using an 8-batch size and 100 epochs, yielding computational results that allow us to assess the model’s accuracy, precision, recall, and dataset classification. In best.pt, the outcome is saved as a weight. It can also be utilized to enhance the model to get better outcomes.When the model has finished being trained, the classification result will be generated.
G. Validation
The generated precision curve, recall curve, precision-recall curve, and confusion matrix could be used to validate the trained data and hence validate the model’s performance metrics. The model must be retrained in order for the performance metrics to be updated to the appropriate outcomes if you wish to improve them.
H. Inference
By uploading the images or using the webcam to capture external images, the trained model needs to be inferred. Using the bounding boxes, class probabilities, and object confidences, the model must be able to identify the external data that has been provided.
I. Output
The model’s output would be a result that could fall into any of the following classes: greening, blackspot, canker, or healthy.
IV. EXPERIMENTAL RESULTS
After the photos are trained, the following categorization outcomes are obtained. This classification report provides a thorough analysis of the model’s performance on each class and how it strikes a balance between recall and precision. Additionally, it displays the amount of instances for every class, indicating an imbalance in the classes.
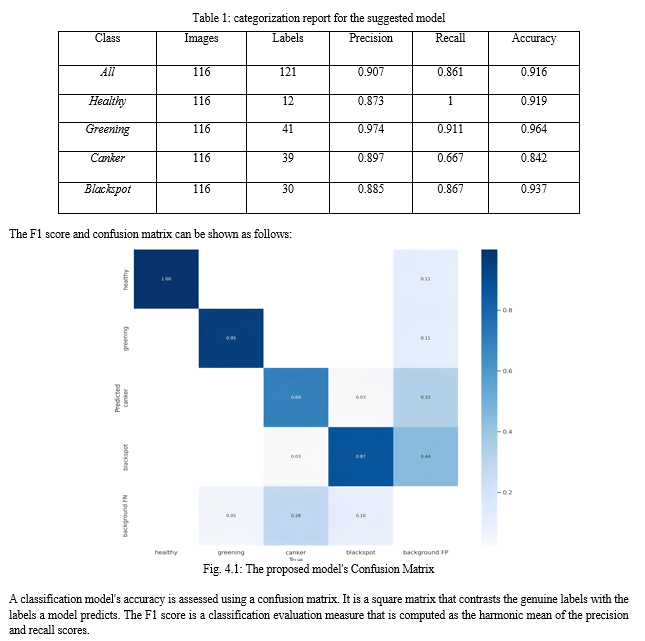
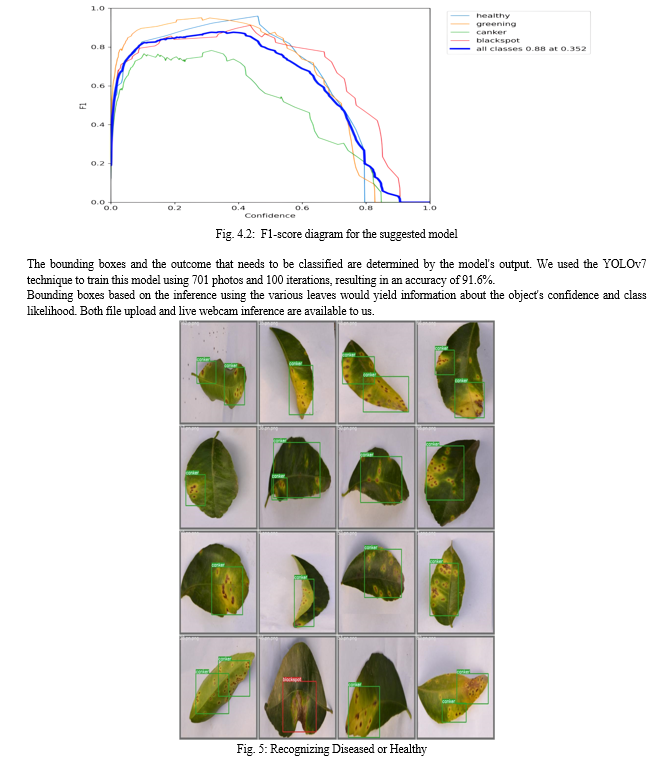
Conclusion
The application of YOLOv7 to the field of plant disease diagnosis offers a strong solution with significant benefits. Through the utilization of the YOLO series\' speed and precision, particularly in its seventh version, this initiative seeks to transform the process of identifying and diagnosing plant illnesses. Deploying such a system in agricultural settings is made more feasible by the architecture\'s real-time capabilities and its capacity to handle high-resolution photos rapidly. The model learns to identify and pinpoint different plant disease kinds through a carefully planned training program, which aids in early identification. In addition to marking a technological advance, the project\'s use of YOLOv7 highlights the technology\'s potential for revolutionary effects on agricultural sustainability and crop yield preservation. The successful application of plant disease diagnosis using YOLOv7 is evidence of the continued synergy between cutting-edge machine learning algorithms and practical agricultural concerns, as the field of computer vision and agriculture continues to expand.
References
[1] Jasim, M. A., & AL-Tuwaijari, J. M. (2020). Plant Leaf Diseases Detection and Classification Using Image Processing and Deep Learning Techniques. In Proceedings of the 2020 International Conference on Computer Science and Software Engineering (CSASE) (pp. 259). Duhok, Kurdistan Region – Iraq. IEEE. [2] P, S., John, A., Devika, V. L., S V, G., & Sakhir, N. (2023). Leaf Disease Detection & Correction using YOLO V7 with GPT3 integrated. International Journal of Engineering Research & Technology (IJERT), Vol. 12(06), IJERTV12IS060064. [3] Deepa, Ms., Rashmi N, & Chinmai Shetty. (Year, if available). A Machine Learning Technique for Identification of Plant Diseases in Leaves. IEEE Xplore Part Number: CFP21F70-ART; ISBN: 978-1-7281-8501-9 [4] Wang, C.-Y., Bochkovskiy, A., & Liao, H.-Y. M. (Year, if available). YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. [5] Akhtar, F., Parthechan, N. A., Daniel, A., Sriramala, S., Mehra, S., & Gupta, N. (Year, if available). Plant Disease Detection based on Deep Learning Approach. 2021 International conference on Advance Computing and Innovative Engineering (ICACITE).
Copyright
Copyright © 2024 Rokkam Sai Praveen, Pothuri Sravani, Savalam Akhil, Tiyyagura Poojithaa Reddy, J M Babu. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET59272
Publish Date : 2024-03-21
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

