Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Pneumonia Detection using Convolutional Neural Network
Authors: Raj Srivastav, Raman Kumar, Shivam Pandey, Ms. Shaba Irram
DOI Link: https://doi.org/10.22214/ijraset.2024.62726
Certificate: View Certificate
Abstract
The deadly illness known as pneumonia develops in the lungs and is brought on by a bacterial or viral infection. Pneumonia can be difficult and prone to error to diagnose in chest X-ray pictures due to its similarities to other lung infections. The purpose of this project is to create a computer-aided pneumonia detection system to speed up the process of making a diagnosis. As a result, an ensemble convolutional neural network (CNN) technique was presented for the automated diagnosis of pediatric pneumonia. In this case, the chest X-ray dataset was used to train seven well-known CNN models (VGG-16, VGG-19, ResNet-50, Inception-V3, Xception, MobileNet, and SqueezeNet) with the proper transfer learning and fine-tuning techniques after they had been pre-trained on the ImageNet dataset. The three best-performing models out of the seven were chosen for the ensemble approach. During the test, the ensemble method was used to combine the predictions made by CNN models to get the final findings. Furthermore, a CNN model was trained from the beginning, and its outcomes were contrasted with those of the suggested ensemble approach.
Introduction
I. INTRODUCTION
The main area of machine learning research in recent years has been computer-aided designs (CAD). Current CAD systems have already demonstrated their ability to help the medical field, particularly with the detection of lung nodules, breast cancer, and mammograms. Not less important are significant features when applying Machine Learning (ML) techniques to medical images. Because of this, the majority of earlier algorithms developed CAD systems by analyzing images using hand-crafted features (Das DK et al., 2013), (Poostchi M et al., 2018). However, there wasn't much useful functionality that could be provided by the hand-crafted features, whose limitations varied depending on the task. The utilization of Deep Learning (DL) models, specifically Convolutional Neural Networks (CNNs), has demonstrated their inherent ability to extract valuable features for image classification tasks (A. S. Razavian et al., 2014). The feature extraction process necessitates the use of transfer learning techniques, in which CNN models that have already been trained on massive datasets like ImageNet are trained on generic features that are then applied to the necessary task. Pre-trained CNN models, such as AlexNet, VGGNet, Xception, ResNet, and DenseNet, are readily available and greatly facilitate the process of extracting significant features. Furthermore, classifications using highly-rich extracted features perform better when it comes to classifying images. In addition to being primarily used to detect lung nodules, chest screening subroutines can also be used to diagnose other conditions like pneumonia, cardiomegaly, effusion, etc. Among these, pneumonia is a contagious and fatal illness that affects millions of people, mostly those over 65 with chronic conditions like diabetes or asthma [5]. Chest X-rays are thought to be the most efficient way to identify the extent and location of the septic region in the lungs during the diagnosis process of pneumonia. Nonetheless, radiotherapists do not take their time reviewing chest radiographs. Pneumonia can appear hazy on chest X-ray images and be mistaken for other conditions.
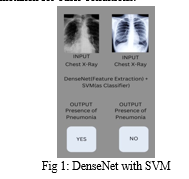
In order to distinguish between abnormal and normal chest X-rays, we assessed the performance of several variations of pre-trained CNN models, which were then followed by various classifiers. The following are the study's major contributions: In order to propose the ideal classifier in the same classification field, (a) a comparative analytical study of various pre-trained CNN models as feature-extractors for analysing chest X-rays is conducted; (b) these models are presented with different classifiers; and (c) the best pre-trained CNN model is evaluated by hyperparameter-tuning the best-analyzed classifier to further improve performance.
This paper's structure is explained as follows: A summary of related research in the same field is provided in Literature review. A description of every detail pertinent to the dataset used is provided in Proposed work. The applied methodology, which has been broken down into several stages, is described in Methods.
II. LITERATURE REVIEW
A. Related Works
One of the most impressive recent developments is the automated diagnosis of pneumonia in chest X-ray images. Deep CNN models have been used in numerous studies to attempt to diagnose pneumonia. A CNN model called CheXNet, with 121 layers, was created by Rajpurkar et al. [6]. Using 100,000 chest X-ray pictures of 14 distinct diseases, they trained CheXNet. Four hundred chest X-ray pictures were used to test the suggested model, and the outcomes were compared to those of radiologists with expertise in the field.
Consequently, it was observed that the CNN model, which is based on deep learning, outperformed radiologists on average in terms of pneumonia detection. Transfer learning was used by Kermany et al. (Kermany DS et al., 2018) to train a CNN model that identified pneumonia in chest X-ray images.
A CNN-based system was used by Rajaraman et al. (Rajaraman S. et al., 2018) to categorize chest X-rays as normal versus viral pneumonia, bacterial versus viral pneumonia, and normal, bacterial versus viral pneumonia.
Using region of interest areas (ROI) containing only the lungs rather than the entire image, they trained CNN models. CNN model was proposed by Stephen et al. (Stephen O et al., 2019). They trained the CNN model from scratch to extract attributes from a given chest X-ray image to achieve remarkable classification performance, and then they used it to determine whether or not a person had pneumonia. This is in contrast to other methods that rely only on transfer learning or traditional handcrafted techniques.
Using a CNN model architecture, residual connections and dilated convolution techniques were used by Liang and Zheng (Liang G et al., 2020) to detect pneumonia.
A sequential CNN model with eighteen layers that performs automatic pneumonia diagnosis was proposed by Siddiqi (Siddiqi, Raheel et al., 2019). Chouhan et al. employed ensemble methodology and transfer learning to detect pneumonia using five CNN models that had already been trained on ImageNet. A two-step procedure to distinguish between viral and bacterial pneumonia was presented by Gu et al. (Gu Xianghong et al., 2018). The suggested approach uses a deep convolutional neural network (DCNN) for pneumonia category classification and a fully convolutional network (FCN) for lung region identification.
Rahman et al. (Rahman Tawsifur et al., 2020) used the transfer learning approach to identify pneumonia using four pre-trained CNN models on ImageNet. Three distinct classification schemes were used to categorize chest radiography images: normal versus viral pneumonia, bacterial versus viral pneumonia, and normal, bacterial versus viral pneumonia.
Three well-known CNN models were employed by Togacar et al. (To?açar et al., 2019) for the feature extraction stage of the pneumonia classification problem. Using the same set of data, they trained each model independently, extracting 1000 features from the final fully connected layers of each CNN. The pneumonia classification problem was solved with 1000 features, which resulted in a reduction in the usage of the minimum redundancy maximum relevance (mRMR) feature selection method and produced features that were fed into machine learning classification algorithms. A multi-layered capsule CapsNet CNN model was proposed by Mittal et al. (Mittal Ansh et al., 2020) to diagnose pneumonia in chest X-ray images.
III. PROPOSED WORK
A. DataSet Description
The dataset used is ChestX-ray14, which comprises 112,120 frontal chest X-ray images from 30,085 patients and was made publicly available on the Kaggle platform by Wang et al. (2017) . The dataset contains radiographic images that are labelled with one or more of the fourteen distinct thoracic diseases. These labels, which were determined by text-mining disease classification from the related radiological reports using Natural Language Processing (NLP), are anticipated to be more than 90% accurate. For the purposes of this work, we treat the labels as ground truth in order to detect pneumonia, in accordance with previous approaches. The largest dataset of chest radiographs that was made available to the public before this one was released was Openi, which included about 4,143 X-ray images.
The resolution of every radiograph image in the dataset is 1024 by 1024. 1431 of these 112,120 photos have been identified as having pneumonia. The dataset has been balanced for binary classification by selecting 1431 normal X-ray images (labelled as "No Findings"). The final dataset utilized for the classification task is made up of 1431 positive image samples (labelled as "Pneumonia") and 1431 negative image samples (labelled as "No Findings"). This subset of the original dataset was used in the process. Subsequently, the dataset was split into two halves, from which 573 randomly selected images were used for testing purposes.
IV. METHODOLOGY
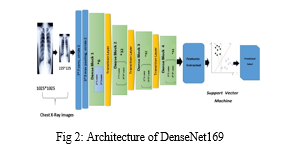
The comprehensive explanation of the used methodology is covered in this section. Figure 2 describes the suggested pneumonia detection system that makes use of the "Densely Connected
Convolutional Neural Network" (DenseNet-169). There are three distinct stages in the architecture of the suggested model: pre-processing, feature extraction, and classification.
A. The Pre-Processing Stage
Reducing the computational complexity of the model, which is likely to increase if the input consists of images, is the main objective of using convolutional neural networks in the majority of image classification tasks. The initial three-channel images were resized from 1024 x 1024 pixels to 224 x 224 pixels in order to expedite processing and lessen computational burden. These resized images have been subjected to every additional technique.
B. The Feature-Extraction Stage
Despite using various variations of pre-trained CNN models to extract the features, the statistical results suggested that DenseNet-169 was the best model for the feature extraction phase. Consequently, the description of the DenseNet-169 model architecture and its role in feature extraction are covered in this step.
- Architecture of DenseNet-169
Because convolutional and pooling layers in deep convolutional networks (DCNNs) have unique types, these networks have emerged as the most productive frameworks for image recognition. However, as the network deepens, the input data or gradient that once passed through the majority of the layers disappears by the time the final layer is reached. DenseNets solve the gradient vanishing issue by directly connecting all of the layers with equal feature sizes. The main reason for employing DenseNet architecture as a feature extractor is the ability to extract more generic features from deeper network layers. The feature extraction procedure has made use of the pre-trained Densely Connected Convolutional Neural Network of 169 layers (DenseNet-169). The variant of this model that we used in our study was trained using the extensive publicly available ImageNet dataset, and it was first proposed by Huang et al. (2016) (Stephen O et al., 2019). Three transition layers, four dense blocks, one convolution and pooling layer at the start make up the DenseNet-169 architecture. The last layer, or the classification layer, is present after these layers. The first convolutional layer uses stride 2 to perform 7×7 convolutions, and then it uses stride 2 to perform a 3×3 max pooling. The network then consists of three sets, each consisting of a transition layer and a dense block after it. The gradient flow throughout the network is improved since the lth layer of the network receives the feature-maps of every layer that came before it. Convolutional neural networks primarily aim to sample feature map sizes down, so the DenseNets architecture is divided into multiple densely connected dense blocks mentioned above. This requires concatenating the feature maps of the preceding layers, which cannot be done unless all the feature maps are of the same sizes.
Transition layers are the layers that lie in between these dense blocks. The network's transition layers are composed of a 1×1 convolutional layer, a batch normalization layer, and a 2×2 average pooling layer with a stride of 2. As previously indicated, there are four dense blocks, with two convolution layers in each. The first layer is one × one in size, and the second is three × three. Six, Twelve, 32, and 32 are the sizes of each of the four dense blocks in the DenseNet169 architecture that were pre trained on ImageNet. The last layer, the classification layer, sits next to it. It completes the global average pooling of 7x7. After that, there is a final fully-connected layer with "softmax" as the activation
2. Extraction of Features
With the exception of the final classification layer, all network layers can be processed using the feature extraction method from the model described in this section 4.2.1. Upon obtaining the final feature representation, a 50176×1 dimension vector was interpreted and subsequently used as an input for various classifiers.
C. The Classification Stage
Various classifiers, including Random Forest and Support Vector Machine, were employed for the classification task following feature extraction. However, it was discovered that using Support Vector Machine as the problem's classifier produced the best results. Therefore, to achieve better results, features taken from DenseNet-169 were combined with an SVM
classifier in the best proposed model. The following is a description of the kernel and parameters used with SVM: Let us consider the following scenario: we have a set of training data: (x1, y1), (x2,y2)...(xn,yn). We need to divide the data into two sets of classes: yi ε (0,1) represents the label class and xi ε Fd is the feature vector. The choice of kernel and parameters has a major impact on SVM performance. The Gaussian "radial basis function" kernel (rbf) was employed (Gu Xianghong et al., 2018). The RBF kernel's gamma and C parameters have a significant impact on SVM performance. The gamma parameter, whose larger value indicates "close," and whose smaller value implies "far," is used intuitively to define the amount of influence that a single training example should have. Thus, the gamma parameter displays the inverse of the influence's radius for the samples that the model used to select as support vectors. Conversely, the C parameter makes up for the training samples' incorrect classification. Whereas a high C seeks to accurately classify every training sample by offering the model exemption to choose additional samples as support vectors, a low C offers a smooth surface.
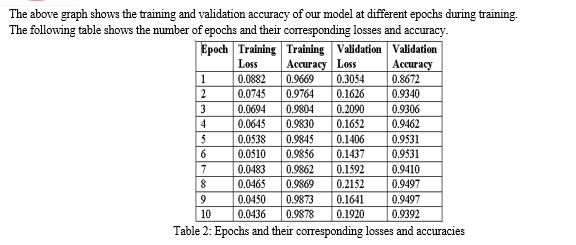
V. LIMITATIONS
Despite the overwhelming results, our model still had certain limitations that we think are important to take into account. The evaluation model's primary limitation is that it does not take the associated patient's history into account. Second, only frontal chest X-rays were employed; however, studies have demonstrated the diagnostic value of lateral view chest X-rays. Thirdly, the model requires a lot of processing power because it uses a lot of convolutional layers; otherwise, computations would take a long time.
Conclusion
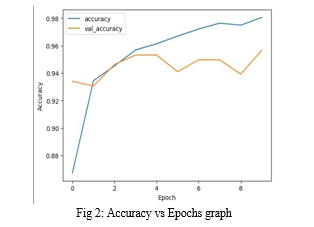
The most important requirement for accurately diagnosing any type of thoracic disease is the presence of skilled radiologists. The main goal of this paper is to increase medical proficiency in places where radiotherapists are still scarce. In such remote areas, our study helps prevent adverse consequences, including death, by facilitating early diagnosis of pneumonia. There hasn\'t been much work done thus far specifically to identify pneumonia in the dataset in question. The creation of algorithms in this field may prove to be very helpful in delivering improved medical care. We evaluated the performance of several pretrained CNN models in this classifier, as well as different classifiers. Based on the statistical findings, we chose DenseNet-169 for the feature extraction stage and SVM for the classification stage. Additionally, we demonstrated how improving the hyper-parameters during the classification stage improved the model\'s performance. Our goal in conducting these experiments is to produce the most dominant pre-trained CNN model and classifier for use in upcoming studies in this field of study. In the near future, improved algorithms for diagnosing pneumonia are probably going to be developed as a result of our study.
References
[1] Das DK, Ghosh M, Pal M, Maiti AK, Chakraborty C. Machine learning approach for automated screening of malaria parasite using light microscopic images. Micron. 2013 Feb;45:97-106. doi: 10.1016/j.micron.2012.11.002. Epub 2012 Nov 16. PMID: 23218914. [2] Poostchi M, Silamut K, Maude RJ, Jaeger S, Thoma G. Image analysis and machine learning for detecting malaria. Transl Res. 2018 Apr;194:36-55. doi: 10.1016/j.trsl.2017.12.004. Epub 2018 Jan 12. PMID: 29360430; PMCID: PMC5840030. [3] Ross NE, Pritchard CJ, Rubin DM, Dusé AG. Automated image processing method for the diagnosis and classification of malaria on thin blood smears. Med Biol Eng Comput. 2006 May;44(5):427-36. doi: 10.1007/s11517-006-0044-2. Epub 2006 Apr 8. PMID: 16937184. [4] A. S. Razavian, H. Azizpour, J. Sullivan and S. Carlsson, \"CNN Features Off-the-Shelf: An Astounding Baseline for Recognition,\" 2014 IEEE Conference on Computer Vision and Pattern Recognition Workshops, Columbus, OH, USA, 2014, pp. 512-519, doi: 10.1109/CVPRW.2014.131. keywords: {Feature extraction;Training;Birds;Visualization;Support vector machines;Vectors;Image recognition}, [5] [online] Available: https://www.cdc.gov/features/pneumonia/index.html. [6] Radiologist-level pneumonia detection on chest x-rays with deep learning (2017). arXiv preprint arXiv:1711.05225 [7] Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, Dong J, Prasadha MK, Pei J, Ting MYL, Zhu J, Li C, Hewett S, Dong J, Ziyar I, Shi A, Zhang R, Zheng L, Hou R, Shi W, Fu X, Duan Y, Huu VAN, Wen C, Zhang ED, Zhang CL, Li O, Wang X, Singer MA, Sun X, Xu J, Tafreshi A, Lewis MA, Xia H, Zhang K. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell. 2018 Feb 22;172(5):1122-1131.e9. doi: 10.1016/j.cell.2018.02.010. PMID: 29474911. [8] Rajaraman, S.; Candemir, S.; Kim, I.; Thoma, G.; Antani, S. Visualization and Interpretation of Convolutional Neural Network Predictions in Detecting Pneumonia in Pediatric Chest Radiographs. Appl. Sci. 2018, 8, 1715. https://doi.org/10.3390/app8101715 [9] Stephen O, Sain M, Maduh UJ, Jeong DU. An Efficient Deep Learning Approach to Pneumonia Classification in Healthcare. J Healthc Eng. 2019 Mar 27;2019:4180949. doi: 10.1155/2019/4180949. PMID: 31049186; PMCID: PMC6458916. [10] Liang G, Zheng L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput Methods Programs Biomed. 2020 Apr;187:104964. doi: 10.1016/j.cmpb.2019.06.023. Epub 2019 Jun 26. PMID: 31262537. [11] Siddiqi, Raheel. (2019). Automated Pneumonia Diagnosis using a Customized Sequential Convolutional Neural Network. 64-70. 10.1145/3342999.3343001. [12] Gu, Xianghong & Pan, Liyan & Liang, Hui-Ying & Yang, Ran. (2018). Classification of Bacterial and Viral Childhood Pneumonia Using Deep Learning in Chest Radiography. ICMIP 2018: Proceedings of the 3rd International Conference on Multimedia and Image Processing. 88-93. 10.1145/3195588.3195597. [13] Rahman, Tawsifur & Chowdhury, Muhammad & Khandakar, Amith & Islam, Khandakar & Islam, Khandaker & Mahbub, Zaid & Kadir, Muhammad & Kashem, Saad. (2020). Transfer Learning with Deep Convolutional Neural Network (CNN) for Pneumonia Detection Using Chest X-Ray. Applied Sciences. 10. 3233. 10.3390/app10093233. [14] To?açar, Mesut & Ergen, Burhan & Cömert, Zafer. (2019). A Deep Feature Learning Model for Pneumonia Detection Applying a Combination of mRMR Feature Selection and Machine Learning Models. IRBM. 10.1016/j.irbm.2019.10.006. [15] Mittal, Ansh & Kumar, Deepika & Mittal, Mamta & Saba, Tanzila & Abunadi, Ibrahim & Rehman, Amjad & Roy, Sudipta. (2020). Detecting Pneumonia Using Convolutions and Dynamic Capsule Routing for Chest X-ray Images. Sensors. 20. 1067-1097. 10.3390/s20041068.
Copyright
Copyright © 2024 Raj Srivastav, Raman Kumar, Shivam Pandey, Ms. Shaba Irram. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET62726
Publish Date : 2024-05-26
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

