Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Chronic Kidney Disease Prediction Using Machine Learning
Authors: Milind Rane, Megha Derkar, Devansh Kabra, Tanuja Desai
DOI Link: https://doi.org/10.22214/ijraset.2024.62593
Certificate: View Certificate
Abstract
Chronic kidney disease (CKD) is a progressive condition in which the kidneys lose their ability to function effectively over time. Individuals with hypertension, diabetes, or a family history of CKD are at increased risk, emphasizing the importance of early detection for effective intervention and management. Recent research has focused on employing machine learning techniques, including Ant Colony Optimization (ACO) and Support Vector Machine (SVM) classifiers, to predict CKD presence using a minimal set of features. This study aims to optimize predictive accuracy through advanced machine learning methodologies, facilitating timely and targeted healthcare interventions for at-risk individuals. By analyzing relevant clinical data, predictive models developed in this research offer promising avenues for early identification of CKD, enabling proactive disease management strategies. The integration of machine learning techniques in CKD prediction not only enhances predictive accuracy but also contributes to advancing personalized healthcare. Early detection allows for the implementation of preventive measures and personalized treatment plans, ultimately improving patient outcomes and reducing the burden on healthcare systems. The proposed methodology seeks to optimize predictive accuracy, aiding in timely and targeted healthcare interventions.
Introduction
I. INTRODUCTION
Chronic Kidney Disease (CKD) represents a significant health challenge on a global scale, affecting millions of people globally and creating significant pressure on healthcare systems.. Detecting CKD early and intervening promptly are crucial for managing the condition effectively and preventing it from advancing to severe stages. Recognizing the urgency of this issue, researchers worldwide are exploring various approaches, including sophisticated machine learning algorithms, to develop predictive models for CKD. The objective is to utilize computational algorithms to analyze
diverse datasets and create reliable tools capable of identifying individuals at an early stage who are at risk of CKD. This empowers healthcare professionals to implement preventive measures, tailor interventions, and ultimately improve patient outcomes. The importance of CKD is highlighted by the challenges associated with its early detection, underlining the potential of machine learning to transform predictive healthcare.
Machine learning, a branch of artificial intelligence, allows computers to learn from data and generate predictions or decisions without being directly programmed for specific tasks. When applied to chronic kidney disease (CKD), machine learning algorithms process diverse datasets that include patient demographics, medical histories, lab test results, and imaging studies. By identifying patterns and associations within this data, these algorithms can pinpoint individuals who may be at an increased risk of developing CKD.
In addition to benefiting patient outcomes, the development of effective CKD prediction models has the potential to positively impact public health and healthcare delivery systems. By advancing the integration of healthcare and machine learning, researchers aim to provide valuable insights derived from computational algorithms and diverse datasets The ultimate goal is to establish pathways for the more personalized and streamlined management of Chronic Kidney Disease, tackling a crucial requirement within global healthcare.
II. LITERATURE SURVEY
[3] Gunarathne, W. H. S. D., K. D. M. Perera, and K A. D. C. P. Kahandawaarachchi : The literature review on kidney disease prediction via data mining techniques reveals varied approaches. SVM outperformed Naïve Bayes in one study, while K-Star and Random Forest showed promise in another. ANN was recommended for dialysis survivability prediction, and individual visit data was deemed crucial for accuracy. The proposed work utilizes SVM, KNN, and Bayes classifiers for Chronic Kidney Disease prediction.
[9] G. Chen et al: Various studies have delved into machine learning applications for kidney disease detection and classification. Ali et al. employed Neighbourhood Component Analysis (NCA) and LSTM to classify subtypes, while Sheehan et al. introduced a Deep Neural Network (DNN) for histologic phenotypes. Ren et al. proposed a Hybrid Neural Network (HNN) utilizing Electronic Health Record (EHR) data, and Santini et al. developed EMS-DLA for tumor segmentation. Additionally, Stefan et al. demonstrated the effectiveness of their AHDCNN model in predicting chronic kidney disease. These studies collectively showcase the potential of diverse datasets and neural network architectures in advancing the diagnosis of kidney diseases.
[10] A. Charleonnan, T. Fufaung, T. Niyomwong, W. Chokchueypattanakit, S. Suwannawach and N. Ninchawee: The research investigates four machine learning techniques to predict chronic kidney disease: K-nearest neighbors, support vector machine, decision tree, and logistic regression. These techniques are evaluated for their classification performance. The methodology includes attribute transformation, feature selection, model building, and evaluation. Using a dataset from the UCI machine learning repository, the experimental results assess metrics such as accuracy, sensitivity, and specificity. The goal of the study is to improve diagnostic efficiency through various computational methods.
[12] U. N. Dulhare and M. Ayesha: Numerous studies investigate data mining methods for predicting Chronic Kidney Disease (CKD) by employing classification algorithms such as K-nearest neighbors, support vector machines, and decision trees. Researchers achieved high accuracy with different classifiers, such as Multilayer Perceptron and Naïve Bayes. However, challenges remain in integrating classification techniques with action rule generation for predicting CKD stages. Proposed methodologies involve data preprocessing, feature selection, and prediction using classifiers like Naïve Bayes. Additionally, predicting CKD stages involves calculating Glomerular Filtration Rate (GFR) and generating action rules based on predicted stages.
[14] Z. Chen, X. Zhang and Z. Zhang: The chronic kidney disease (CKD) dataset, obtained from the UCI Machine Learning Repository, includes 400 instances with 14 numerical and 10 categorical attributes, plus a class label. Post-preprocessing, 386 instances with 21 features were retained. To mimic environmental noise, varying levels of disturbances were introduced, creating three different composite datasets. Numerical attributes were normalized to the range [0, 1]. Multivariate models such as K-nearest neighbor (KNN), support vector machine (SVM), and soft independent modeling of class analogy (SIMCA) were developed and assessed using bootstrapped Latin partition cross-validation in MATLAB..
[16] K. A. Padmanaban and G. Parthiban: The study employs data mining techniques, including classification and evaluation methods, to forecast Chronic Kidney Disease (CKD) risk. It utilizes machine learning algorithms and tools like RapidMiner, WEKA, and YALE. The architecture incorporates 10-fold cross-validation for enhanced performance. Attributes such as sex, age, weight, smoking, and blood test results are considered. Naïve Bayes and Decision Tree methods are used for classification, assessing model accuracy and risk.
[18] M. S. Basarslan and F. Kayaalp: The study explores attribute selection methods, including Correlation-Based Attribute Selection (CBAS) and Fuzzy Rough Set-Based Attribute Selection (FRSBAS), for Chronic Kidney Disease (CKD) detection. It employs four classification algorithms: k-Nearest Neighbor, Naive Bayes, Logistic Regression, and Random Forest. Performance evaluation metrics like accuracy, precision, and sensitivity are employed to gauge model effectiveness, with FRSBAS consistently showcasing superior outcomes.
[19] R. Devika, S. V. Avilala, and V. Subramaniyaswamy: The literature review compares Naive Bayes, K-Nearest Neighbors (KNN), and Random Forest classifiers for predicting chronic kidney disease. Various studies suggest the effectiveness of these algorithms in medical diagnosis due to their ability to handle diverse data types and complexities. However, their performance varies based on factors such as dataset characteristics, feature selection, and preprocessing techniques.
[27] P. Yildirim: The text discusses the class imbalance problem in data mining and machine learning, particularly in medical datasets, and explores sampling methods such as under sampling and over sampling. It then delves into the multilayer perceptron (MLP) neural network architecture, its training using back propagation, and related research on MLP-based decision support systems for medical diagnosis. Finally, it presents experimental results comparing sampling methods' performance in predicting chronic kidney disease using MLPs.
III. METHODOLOGY
A. Dataset
The dataset is sourced from the publicly available Chronic Kidney Disease (CKD) Dataset provided by the UCI repository. Comprising 400 samples categorized into two distinct classes, the dataset encompasses 25 attributes. Among these attributes, 11 are of a numeric nature, 13 are nominal, and one is designated as the class attribute.
It is worth noting that the dataset includes instances with missing values. The dataset draws on patient data, incorporating information such as age, blood pressure, specific gravity, albumin, sugar, red blood cells, among other relevant attributes. This diverse set of features enables a comprehensive analysis of factors contributing to chronic kidney disease. The utilization of a publicly accessible dataset ensures transparency and facilitates the reproducibility of research findings
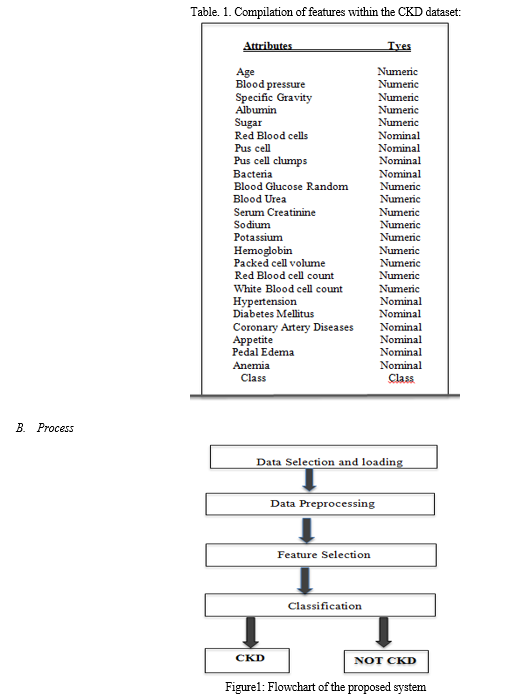
- Data Preprocessing
Data pre-processing refers to the initial stage of data analysis where raw data is cleaned, transformed, and prepared for further analysis. It's the process of cleaning and organizing raw data into a usable format for further analysis. In datasets characterized by features, handling missing values becomes crucial. Numeric values are transformed into 'float64,' while categorical nulls are typically filled with the most frequent value. Label encoding is then employed to convert categorical attributes into 'int' types. The 'imputer' function is utilized to calculate means for each column, which are then used to replace missing values. This meticulous process ensures that the dataset is standardized and ready for thorough analysis using machine learning techniques.
Following data pre-processing, the dataset undergoes training, validation, and testing phases. During the training phase, algorithms are trained to build models based on the dataset. Validation is employed to assess or enhance the fits of these models, ensuring their effectiveness and reliability. In the final testing phase, the model hypothesis undergoes evaluation, offering valuable insights into how well it performs and its ability to make predictions.
2. Feature Selection
Feature selection is a technique used in machine learning to identify and select the most relevant features that significantly contribute to predicting variables or outcomes. Ant Colony Optimization (ACO) in this study is employed as a sophisticated technique for efficient feature selection. This method draws inspiration from the behaviour of real ants, which navigate through complex environments to find optimal paths.
In the context of feature selection, ACO operates similarly to real ants by exploring various feature subsets and evaluating their performance. Artificial Ants represent a multi-agent approach, where each ant corresponds to a potential solution consisting of a subset of features. These ants communicate through pheromones, mimicking how real ants leave chemical trails to guide their fellow colony members.
During iterations of the ACO algorithm, ants construct solutions by probabilistically selecting features based on pheromone levels and heuristic information. Pheromone intensity reflects the quality of feature subsets, with higher intensity indicating better performance. Ants with successful feature subsets deposit more pheromones, influencing subsequent iterations to favor those features.
The process of feature selection using ACO involves iteratively evaluating the performance of classification algorithms on different feature subsets. This is achieved through a wrapper evaluation function, which assesses the predictive accuracy or other performance metrics of the model. By iteratively refining feature subsets based on the feedback from classification algorithms, ACO effectively identifies the most informative features for predictive modelling.
3. Classification using Random Forest:
In the classification phase , we utilize the Random Forest algorithm as the predictive model for disease outcomes, opting for the scikit-learn library's Random Forest Classifier over Support Vector Machine (SVM). The dataset is partitioned into two distinct sets: one designated for training purposes and the other for testing. The Random Forest model is trained solely on the training data. Predictions are subsequently generated using the 'predict' method from the Random Forest Classifier.
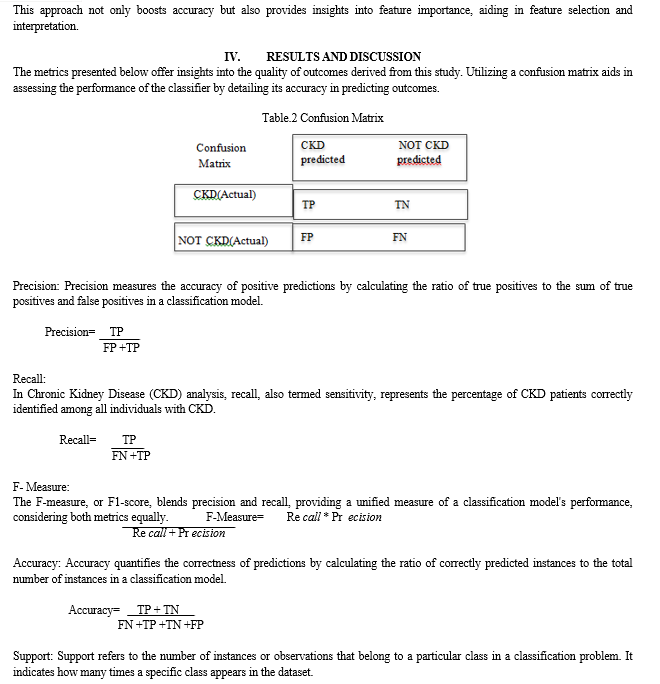
To comprehensively assess the model's performance, we utilize a confusion matrix, which breaks down true positives, true negatives, false negatives and false positives. This evaluation helps measure accuracy, recall, precision and F1-score.By scrutinizing these metrics, we gain insights into the model's efficacy in correctly classifying disease outcomes, thus informing further refinements or adjustments to enhance its predictive capabilities.
4. Random Forest in Machine Learning:
Random Forest builds multiple decision tree using random subsets of data and features, providing robust predictions and feature importance. Unlike Support Vector Machine (SVM), it embraces an ensemble learning approach, which involves building numerous decision trees during training and combining their outputs to enhance accuracy and resilience.
In the realm of binary linear classification, Random Forest excels by creating an ensemble of decision trees, where each tree plays a role in the final prediction. This ensemble nature underscores its adaptability and effectiveness across a spectrum of applications. By harnessing insights from multiple trees, Random Forest can grasp intricate data relationships, yielding resilient predictions less prone to overfitting or noise.
This approach not only boosts accuracy but also provides insights into feature importance, aiding in feature selection and interpretation.
IV. RESULTS AND DISCUSSION
The metrics presented below offer insights into the quality of outcomes derived from this study. Utilizing a confusion matrix aids in assessing the performance of the classifier by detailing its accuracy in predicting outcomes.
Conclusion
By employing Ant Colony Optimization (ACO) for feature selection and the Support Vector Machine (SVM) algorithm for classification, we attained precise predictions with minimal attributes used. This strategic approach enables us to efficiently identify the most relevant features, optimizing the overall performance of our predictive model. Notably, this methodology showcases its efficacy not only in our specific domain but also across a wide array of applications and industries. By leveraging advanced optimization techniques and robust classification algorithms, we demonstrate the potential for significant advancements in predictive modelling, thereby enhancing decision-making processes and driving impactful outcomes across various domains.
References
[1] Sinha, Parul, and Poonam Sinha. \"Comparative study of chronic kidney disease prediction using KNN and SVM.\" International Journal of Engineering Research and Technology 4, no. 12 (2015): 608-12. [2] Yildirim, Pinar. \"Chronic Kidney Disease Prediction on Imbalanced Data by Multilayer Perceptron: Chronic Kidney Disease Prediction.\" In Computer Software and Applications Conference (COMPSAC), 2017 IEEE 41st Annual, vol.3(2008):095-2. [3] Gunarathne, W. H. S. D., K. D. M. Perera, and K A. D. C. P. Kahandawaarachchi. \"Performance Evaluation on Machine Learning Classification and Data Analytics for Chronic Kidney Disease (CKD).\" In Bioinformatics and Bioengineering (BIBE), 2017 IEEE 17th International Conference on, pp. 291-296. IEEE, 2017. [4] Tafadzwa L. Chaunzwa, Ahmed Hosny, Yiwen Xu, Andrea Shafer, Nancy Diao “Deep learning classification of Chronic kidney diseases”. [5] Venkata Tulasiramu Ponnada, S.V. Naga Srinivasu “Efficient CNN for Kidney Disease Detection” [6] Wafaa Alakwaa,Mohammad Nassef,Amr “Kidney Cancer Detection and Classification with 3D Convolutional Neural Network (3D-CNN)” [7] Abdulrazak Yahya Saleh ,Chee Ka Chin,Vanessa Penshie ,Hamada Rasheed Hassan “Kidney cancer medical images classification using hybrid CNN-SVM” [8] P. Arulanthu and E. Perumal, \"Predicting the Chronic Kidney Disease using Various Classifiers\", 2019 4th International Conference on Electrical Electronics Communication Computer Technologies and Optimization Techniques (ICEECCOT), pp. 70-75, Dec. 2019. [9] G. Chen et al., \"Prediction of Chronic Kidney Disease Using Adaptive Hybridized Deep Convolutional Neural Network on the Internet of Medical Things Platform\", IEEE Access, vol. 8, pp. 100497-100508, 2020. [10] A. Charleonnan, T. Fufaung, T. Niyomwong, W. Chokchueypattanakit, S. Suwannawach and N. Ninchawee, \"Predictive analytics for chronic kidney disease using machine learning techniques\", 2016 Management and Innovation Technology International Conference (MITicon), pp. MIT-80-MIT-83, Oct. 2016 [11] .Siddeshwar Tekale, “Prediction of Chronic Kidney Disease Using Machine Learning, International Journal of Advanced Research in Computer and Communication Engineering, 2018. [12] U. N. Dulhare and M. Ayesha, \"Extraction of action rules for chronic kidney disease using Naïve bayes classifier,\" 2016 IEEE International Conference on Computational Intelligence and Computing Research (ICCIC), Chennai, 2016, pp. 1-5. [13] A. J. Aljaaf et al, \"Early prediction of chronic kidney disease using machine learning supported by predictive analytics,\" in 2018 IEEE Congress on Evolutionary Computation (CEC), 2018, . [14] Z. Chen, X. Zhang and Z. Zhang, \"Clinical risk assessment of patients with chronic kidney disease by using clinical data and multivariate models,\" Int. Urol. Nephrol., vol. 48, (12), pp. 2069-2075, 2016. [15] Baisakhi Chakraborty, “Development of Chronic Kidney Disease Prediction Using Machine Learning”, International Conference on Intelligent Data Communication Technologies, 2019. [16] K. A. Padmanaban and G. Parthiban, \"Applying Machine Learning Techniques for Predicting the Risk of Chronic Kidney Disease,\" Indian Journal of Science and Technology, vol. 9, (29), 2016. [17] S. K. Dowluru and A. K. Rayavarapu, “Statistical and data mining aspects on kidney stones: a systematic review and metza-analysis,” Open Access Scientific Reports, vol. 1, no. 12, 2012. [18] M. S. Basarslan and F. Kayaalp, “Performance analysis of fuzzy rough set-based and correlation-based attribute selection methods on detection of chronic kidney disease with various classifiers,” in Proceedings of the 2019 Scientific Meeting on Electrical-Electronics and Biomedical Engineering and Computer Science (EBBT), IEEE, Istanbul, Turkey, April 2019. [19] R. Devika, S. V. Avilala, and V. Subramaniyaswamy, “Comparative study of classifier for chronic kidney disease prediction using naive Bayes, KNN and random forest,” in Proceedings of the 2019 3rd International Conference on Computing Methodologies and Communication (ICCMC), Erode, India, March 2019. [20] S. Khan, M. Z. Khan, P. Khan, G. Mehmood, A. Khan, and M. Fayaz, “An ant-hocnet routing protocol based on optimized fuzzy logic for swarm of UAVs in FANET,” Wireless Communications and Mobile Computing, vol. 2022, Article ID 6783777, 12 pages, 2022. [21] M. Z. U. Haq, M. Z. Khan, H. U. Rehman et al., “An adaptive topology management scheme to maintain network connectivity in Wireless Sensor Networks,” Sensors, vol. 22, no. 8, p. 2855, 2022. [22] F. E. Murtagh, J. Addington-Hall, P. Edmonds, P. Donohoe, I. Carey, K. Jenkins, et al., \"Symptoms in the month before death for stage 5 chronic kidney disease patients managed without dialysis\", Journal of pain and symptom management, vol. 40, no. 3, pp. 342-352, 2010. [23] D. Dua and C. Graff, \"UCI machine learning repository\", 2017, [online] Available: http://archive.ics.uci.edu/ml. [24] W. Gunarathne, K. Perera and K. Kahandawaarachchi, \"Performance evaluation on machine learning classification techniques for disease classification and forecasting through data analytics for chronic kidney disease (ckd)\", 2017 IEEE 17th International Conference on Bioinformatics and Bioengineering (BIBE). IEEE, pp. 291-296, 2017. [25] E.-H. A. Rady and A. S. Anwar, \"Prediction of kidney disease stages using data mining algorithms\", Informatics in Medicine Unlocked, vol. 15, pp. 100178, 2019. [26] C. Li, \"Little’s test of missing completely at random\", The Stata Journal, vol. 13, no. 4, pp. 795-809, 2013. [27] P. Yildirim, \"Chronic kidney disease prediction on imbalanced data by multilayer perceptron: Chronic kidney disease prediction\", 2017 IEEE 41st Annual Computer Software and Applications Conference (COMPSAC), vol. 02, pp. 193-198, 2017.
Copyright
Copyright © 2024 Milind Rane, Megha Derkar, Devansh Kabra, Tanuja Desai. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET62593
Publish Date : 2024-05-23
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

