Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- References
- Copyright
Plant Disease Detection
Authors: Barnaba Vanlalhruaizela, Lalremsangi , Loicy Lalrinnungi, Pragnya N, Vinutha M S
DOI Link: https://doi.org/10.22214/ijraset.2024.62750
Certificate: View Certificate
Abstract
This project addresses the critical challenge of plant disease detection in India, a nation where agriculture supports nearly 70% of the population. Traditional procedures for identifying plant diseases are both labor-intensive and time-consuming. To enhance efficiency and accuracy, this project introduces an innovative approach that integrates machine learning and image processing techniques. By analyzing images of leaves, the system identifies diseases based on key factors such as leaf color, damage extent, area, and texture parameters. Unlike conventional methods that depend on visual inspections or chemical processes, this automated system provides a cost-effective and efficient solution for monitoring extensive crop fields. By leveraging statistical machine learning and advanced image processing algorithms, the proposed solution ensures precise disease detection with minimized computational complexity and reduced prediction time.
Introduction
I. INTRODUCTION
Farming shapes the spine of India's economy, with around 70% of its populace depending on this segment. Be that as it may, guaranteeing a solid surrender in the midst of different challenges, counting plant maladies, remains a basic concern. Recognizing plant illnesses expeditiously is crucial to checking misfortunes in agrarian efficiency. Manual perception of plant illnesses demonstrates to be difficult, requiring critical labor, ability, and time. To address these challenges, the integration of picture preparing and machine learning models develops as a promising arrangement for infection location. This venture points to investigate methods for successfully identifying plant illnesses through the examination of leaf images.
Image handling, a specialized field inside flag preparing, plays a significant part in extricating germane data from pictures. Complementing this, machine learning, a subset of counterfeit insights, robotizes errands by learning from information. By leveraging machine learning calculations, we point to observe designs inside preparing information, encouraging precise infection location. Parameters such as leaf color, degree of harm, leaf range, and surface are urgent for classification. Through comprehensive investigation of these picture parameters, our objective is to accomplish ideal exactness in malady identification.
Traditionally, plant illness discovery depended on manual visual assessment by specialists or included chemical forms, both of which are labor-intensive and exorbitant, particularly for large-scale cultivating operations. In differentiate, the proposed arrangement offers a more productive and cost-effective approach. By leveraging robotized infection location based on leaf indications, our framework empowers opportune intercession and diminishes the require for broad human observing. In addition, the computational effectiveness of our approach, utilizing measurable machine learning and picture handling calculations, improves its common sense and versatility for real-world rural applications.
II. PROBLEM STATEMENT
A. Literature Survey
[1] Jianping Yao1 · Son N. Tran1,3 · Samantha Sawyer2 · Saurabh Garg1 - Machine learning techniques for categorizing leaf diseases can be grouped into three main categories: shallow learning (SL), deep learning (DL), and augmented learning (AL). Shallow learning involves first locating the leaf and extracting features before categorizing the disease. Deep learning, in contrast, uses an end-to-end process that bypasses feature engineering by directly learning classifiers from input images. Due to its efficiency, deep learning has shown considerable potential in this area. However, the comparative advantages and disadvantages of shallow and deep learning methods are still being actively researched.
[2] Yasmin M. Alsakar , Nehal A. Sakr , and Mohammed Elmogy - Color is a fundamental attribute for plant image representation, widely used in image retrieval due to its invariance to translation, scale, and rotation. Color feature extraction encompasses aspects like as color space, similarity measurements, and color quantization. Numerous color descriptors, including color histograms, play a significant role in this process. Singh et al. introduced a color slicing method specifically for detecting paddy blast disease, converting RGB to HSI and employing color slicing for diseased area extraction. This approach outperformed canny and Sobel methods, achieving an accuracy of 96.6%.
[3] Shoaib M, Shah B, EI-Sappagh S, Ali A, Ullah A, Alenezi F, Gechev T, Hussain T and Ali F - The application of machine learning (ML) and deep learning (DL) techniques in plant disease detection has demonstrated significant potential in identifying plant diseases through digital images with high accuracy. Traditional ML approaches, including feature extraction and classification, utilize image characteristics such as texture, shape, and color to train categorizers that distinguish between healthy and diseased plants. These methods have been successful in detecting diseases like leaf blotch, powdery mildew, and rust, as well as symptoms from abiotic stresses such as drought and nutrient deficiency. However, they repeatedly fall short in detecting early-stage diseases and processing complex, high-resolution images. Recent advancements incorporate more sophisticated techniques to overcome these limitations, aiming to increase the accuracy and robustness of plant disease detection systems.
B. Objectives
The project focuses on a critical issue in India's agricultural sector: detecting plant diseases. With approximately 70% of the population reliant on agriculture, prompt identification of diseases is essential to reduce crop losses. This initiative seeks to create an effective and accessible solution using image processing and machine learning technologies. The specific goals are:
- Developing a robust image processing framework that can extract essential features from leaf images to aid in disease classification.
- Utilizing machine learning algorithms to analyze the image data and accurately identify various plant diseases based on parameters such as leaf color, damage extent, area, and texture.
- Streamlining the detection process to reduce the need for manual observation or chemical methods, offering a cost-effective and time-efficient alternative for monitoring extensive crop fields.
C. Existing System
In the current framework, different components are utilized to screen and oversee agrarian maladies successfully. Sensor systems are sent in areas to track natural parameters like stickiness, temperature, and soil dampness, with deviations showing potential malady or bother nearness. Climate information integration helps in foreseeing infection episodes by analyzing climate designs, encouraging the usage of preventive measures. Infection models, utilizing chronicled information, estimate malady event and spread considering components like plant assortments and neighborhood climate. Coordinates Bother Administration (IPM) procedures are executed, combining natural, social, and chemical control strategies to moderate illness affect whereas advancing feasible horticulture. Moreover, government-led activities in certain nations back malady checking and administration endeavors, advertising assets and help to ranchers.
D. Proposed System
The proposed framework integrates Calculated Relapse, Back Vector Instrument (SVM), and Support Learning to enhance both double and multiclass classification in forecasting plant diseases. By leveraging Calculated Regression's simplicity and efficiency, SVM's ability to identify optimal hyperplanes, and Support Learning's sequential decision-making capability, the framework ensures accurate and timely disease control interventions. Additionally, on-site field access facilitates authentic data collection and validation, while a dedicated office space equipped with essential resources fosters collaborative improvement. Reliable internet connectivity promotes seamless access to online resources and cloud-based services, enhancing the framework's effectiveness and relevance in real-world agricultural settings.
III. EXPERIMENT
In this project, both Jupyter Notebook and Visual Studio Code (VS Code) function as integrated development environments (IDEs), offering versatile platforms for code development, experimentation, and documentation. Jupyter Notebook facilitates interactive coding and visualization, making it particularly valuable for prototyping and exploring data-driven solutions. On the contrary, Visual Studio Code provides a comprehensive development environment with robust support for web development tools, machine learning frameworks, and seamless integration with version control systems.
The dataset employed for training and testing purposes is the TIMIT dataset, comprising audio recordings paired with corresponding phonetic annotations. To support the system's requirements, high-performance hardware with GPU support is essential, alongside software components such as Python, TensorFlow, and associated libraries.
Model training entails constructing a Bidirectional LSTM neural network supplemented with dropout layers, with the objective of classifying phonetic units based on audio features. Upon training, the model demonstrates an accuracy of approximately 73% on phonetic classification tasks. Additionally, visualization techniques, such as confusion matrices, are utilized to assess the model's performance across diverse phonetic categories.
Lastly, the model's functionality extends to translating phonetic units into corresponding viseme classifications, offering valuable insights into the mapping between speech and facial expressions. This capability contributes to a deeper understanding of speech-to-facial expression dynamics, enhancing applications in various fields such as human-computer interaction and multimedia processing
IV. METHODOLOGY
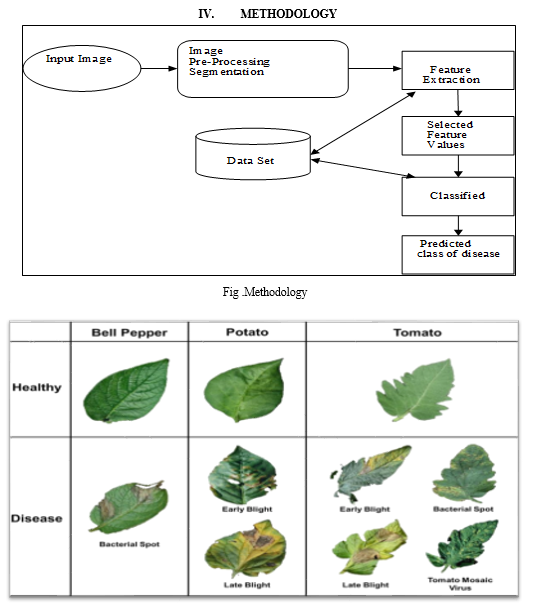
The dataset used in my project consists of RGB images of diseased and healthy crop leaves. It is categorized into various classes representing different types of diseases affecting plants. The dataset is divided into training, validation, and test sets, with a total of approximately 87,000 images. Each image is labeled with the corresponding class, allowing the model to learn the characteristics of different diseases. Dataset used is https://www.kaggle.com/datasets/vipoooool/new-plant-diseases-dataset
In the plant disease recognition system outlined above, a Convolutional Neural Network (CNN) is employed to classify images of plant leaves into various disease categories. CNNs are well-suited for image categorization tasks due to their capacity to learn spatial hierarchies of features automatically and adaptively from input images. The model's accuracy, which is a key performance metric, indicates the proportion of correct predictions it makes out of all predictions. High accuracy signifies that the model is effective in correctly identifying healthy and diseased plants. To further refine the model's performance, data augmentation methods are used to artificially expand the training dataset by making various transformations to the images, thereby improving the model's generalization capabilities. The accuracy of this CNN model, along with other metrics like recall, precision, and F1 score, can be evaluated up to 95% using a validation set, which consists of images not seen by the model during training. This ensures that the model's performance is robust and reliable when deployed in real-world scenarios for early and accurate detection of plant diseases.
Before training the model, we have performed several preprocessing steps on the images. This included resizing the images to a consistent size (e.g., 128x128 pixels) to ensure uniformity, converting the color space from BGR to RGB, and normalizing the pixel values to a range between 0 and 1. Additionally, we used techniques such as data augmentation to improve the diversity of the training data and improve the generalization ability of the model.
For our research project, we will employ a comprehensive methodology integrating various techniques tailored to address the complexities of plant disease prediction in agricultural settings. Firstly, logistic regression will serve as a foundational method, leveraging its simplicity and efficiency for binary classification tasks. This linear model will be adept at predicting the probability of instances belonging to specific classes, providing valuable intuition into disease presence. Additionally, Support Vector Mechanism (SVM) will be utilized to effectively handle both binary and multiclass classification challenges. By identifying hyperplanes that optimally separate classes within a high-dimensional space, SVM offers robust classification capabilities crucial for our predictive model's accuracy. Moreover, we will explore the applicability of reinforcement learning, particularly in scenarios requiring sequential decision-making, such as optimizing disease control interventions. While less commonly employed in plant disease prediction compared to traditional supervised learning methods, reinforcement learning presents promising avenues for enhancing predictive accuracy.
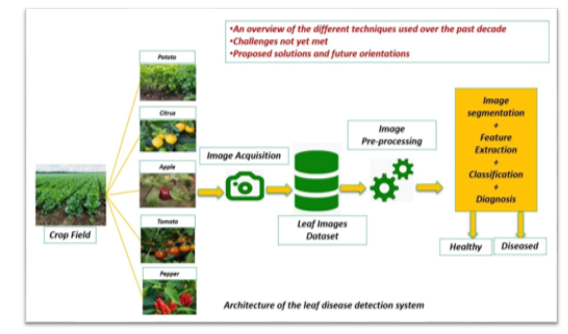
Field access will be paramount, facilitating on-site data collection and validation to ensure the system's development and testing under authentic agricultural conditions. This approach enhances the model's applicability and effectiveness in real-world scenarios. Furthermore, providing a dedicated office space equipped with essential resources for the development team fosters collaborative work, discussions, and project planning. This space will feature workstations, computational resources, and communication tools, optimizing team efficiency and productivity. Lastly, ensuring reliable internet connectivity is imperative, enabling seamless access to online resources, software updates, and potential deployment of the predictive system. A stable and high-speed internet connection facilitates efficient collaboration and access to cloud-based services, augmenting the research endeavor's effectiveness.
V. RESULTS AND DISCUSSION
The implemented system presents a user-friendly interface allowing users to upload leaf images for disease detection. Leveraging image processing techniques, the system accurately identifies spots indicative of diseases on the leaf surface. Moreover, users can utilize the classification feature to determine the specific disease affecting the leaf based on its visual characteristics. By incorporating a treatment page, the system not only diagnoses leaf diseases but also provides actionable information on preventive measures and treatments. This holistic approach empowers users with valuable insights into plant health management, facilitating timely interventions to mitigate disease spread and preserve crop yield. Overall, the system demonstrates promise in enhancing agricultural practices through accessible and informative disease diagnosis and management.
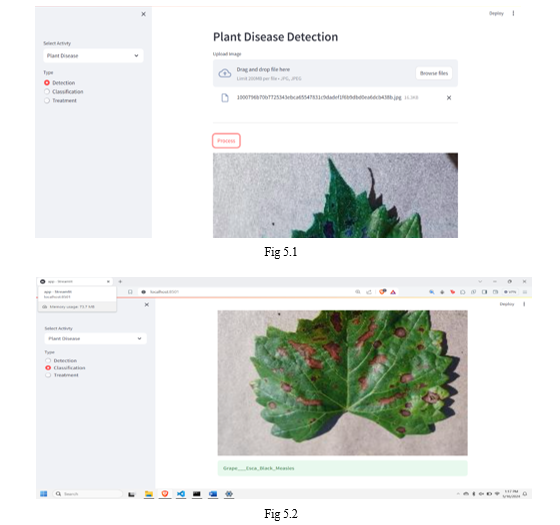
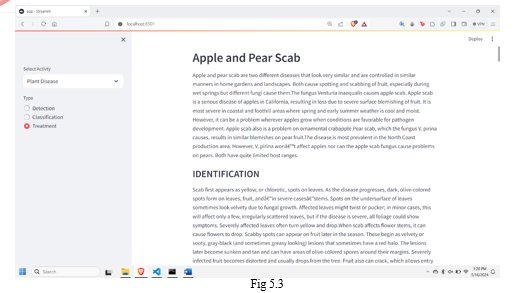
In summary, the plant disease forecasting system represents a significant leap in agricultural innovation, offering farmers a powerful tool to effectively combat crop diseases. By leveraging machine learning, especially Convolutional Neural Networks (CNNs), the system enables early detection and provides valuable insights through thorough data analysis, encompassing both images and environmental factors. Its user-friendly interface ensures accessibility, allowing for seamless image uploads, real-time predictions, and access to pertinent information. Through timely alerts and recommendations, alongside integration into existing agricultural systems, the framework enhances efficiency and usefulness. Moreover, its adherence to ethical considerations, regulatory compliance, user education, and continuous monitoring highlights a comprehensive approach to precision farming. Ultimately, this endeavor signifies a collaborative effort among data scientists, agricultural experts, and technology enthusiasts, marking a significant stride towards sustainable agriculture and enduring positive impact within farming communities.
VI. ACKNOWLEDGMENT
We extend our heartfelt appreciation to Prof. Vinutha M S., our esteemed guide and mentor, for her invaluable unwavering support and guidance throughout the duration of this project. Her expertise in the field of machine learning has been instrumental in shaping the direction and success of our endeavour. Additionally, we express our sincere gratitude to all those who contributed to the realization of our project, from colleagues who offered insightful feedback to collaborators who provided technical assistance. Special recognition is extended to our project supervisor, whose continuous guidance ensured that our efforts remained focused and productive. Furthermore, we extend immense thanks to our dedicated team members whose diligent work and collaboration facilitated the seamless integration of various components, from developing the machine learning model to implementing the end-to-end web application.
References
[1] Machine learning for leaf disease classification: data, techniques and applications. Jianping Yao1 · Son N. Tran1,3 · Samantha Sawyer2 · Saurabh Garg1.Published online: 18 October 2023 [2] Plant Disease Detection and Classi?cation Using Machine Learning and Deep- Conference Paper · January 2023. Yasmin M. Alsakar , Nehal A. Sakr , and Mohammed Elmogy See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/375660975 [3] An advanced deep learning models-based plant disease detection: A review of recent research- Shoaib M, Shah B, EI-Sappagh S, Ali A, Ullah A, Alenezi F, Gechev T, Hussain T and Ali F (2023) An advanced deep learning models-based plant disease detection: [4] S. Khirade et Al, plant disease detection using digital image processing techniques and BPNN(2023). (https://scholar.google.co.in/scholar?hl=en&as_ sdt=0,5&as_vis=1&qsp=2&q=image+processing+plant+disease+detection&qst=br#d=gs_qabs&t=1704178802317&u=%23p%3Dg8xXCqIRdJcJ) [5] Sharath D.M et Al, Bacterial Blight detection system(2021) (https://www.researchgate.net/publication/371755394_A_Systematic_Literature_Review_on _Plant_Disease_Detection_Motivations_Classification _Techniques_Datasets_Challenges_and_Future_Trends) [6] Peyman Moghadam et Al,hyperspectral imaging in plant disease detection and VNIR(2021) (https://www.researchgate.net/profile/Vaishnavi-Monigari/publication/353336310_Plant_Leaf_Disease_Prediction/links/60ffc5442bf3553b29148ded/Plant-Leaf-Disease-Prediction.pdf)
Copyright
Copyright © 2024 Barnaba Vanlalhruaizela, Lalremsangi , Loicy Lalrinnungi, Pragnya N, Vinutha M S. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET62750
Publish Date : 2024-05-26
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

