Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Tomato Leaves Disease Detection using Hybrid Model CNN-LSTM
Authors: Dr. V. Rama Chandran, M Jyothika, M Nandhini, P. Neelima, P Neeraja
DOI Link: https://doi.org/10.22214/ijraset.2024.59173
Certificate: View Certificate
Abstract
This paper delves into the exploration and comparison of Convolutional Neural Networks (CNN), Long Short-Term Memory (LSTM), and a hybrid CNN-LSTM model for the task of tomato leaf disease detection. Through rigorous experimentation and evaluation, it was determined that the CNN-LSTM hybrid model demonstrates superior performance both CNN and LSTM individually in terms of accuracy and robustness. Leveraging a comprehensive dataset comprising images of healthy and diseased tomato leaves, including various disease types, such as Bacterial Spot, Early Blight, Septorial Leaf Spot, Leaf Mold, and Yellow Leaf Curl Virus, the study demonistrate the efficacy of the suggested hybrid approach. The hybrid model capitalizes on the strengths of both CNN and LSTM, integrating spatial and temporal information for enhanced disease identification accuracy. The results highlight the effect of hybrid deep learning structures in agricultural disease management, offering a promising solution for early detection and mitigation of plant diseases. This paper dds to the expanding collection of literature on deep learning applications in agriculture and offers valuable insights for researchers and practitioners seeking to address challenges in crop health monitoring and management.
Introduction
I. INTRODUCTION
Tomato cultivation is a critical component of global agriculture, serving as a key nutritional and economic resource for millions of farmers and consumers around the world. The vitality of tomato plants, however, is under constant siege from a variety of diseases. These afflictions can drastically reduce yields if they are not identified and managed swiftly. The advent of artificial intelligence (AI) and deep learning in past few years has introduced groundbreaking methods for addressing agricultural challenges, particularly in the early detection of plant diseases. Understanding the specific diseases that threaten tomato plants and recognizing their symptoms can greatly aid in their management. The five most common diseases impacting tomato harvests that are focused in the project, along with their distinguishing features:
- Bacterial Spot (caused by the Xanthomonas bacterial pathogen) is identifiable by black-green spots across the foliage, presenting as small dots less than 1/8 inch in diameter. This disease can severely impact the photosynthetic capability of the plants, leading to reduced growth and yield.
- Early Blight (caused by the Alternaria linariae fungus) manifests as brownish spots on the foliage, typically affecting younger leaves first. This disease can lead to significant defoliation, exposing fruits to sunscald and further reducing crop yield.
- Septoria Leaf Spot (caused by the Septoria lycopersici fungus) features yellow to dark brown spots and primarily appears on older leaves. If left unchecked, it can lead to widespread defoliation, stressing the plant and diminishing fruit production.
- Leaf Mold (caused by an Ascomycetes fungal pathogen) results in foliage turning pale and eventually purple. It primarily affects older leaves, which dry up and drop off. This disease thrives in high humidity environments and can spread rapidly if conditions are favourable.
- Yellow Leaf Curl Virus is a virus transmitted by the silverleaf whitefly, causing leaves to curl towards the center. This disease stunts the growth of tomato plants, leading to reduced yields and compromised fruit quality.
Deep learning in agriculture offers a promising solution to disease detection in tomato cultivation. Through the analysis of images captured in the field, algorithms can identify the early signs of these diseases with remarkable accuracy. This capability aids farmers to take proactive actions to control the proliferation of diseases, such as applying targeted treatments or removing infected plants, thereby safeguarding their crops. By harnessing these technologies, farmers can enhance their disease management practices, making them more enhanced and superior in effectiveness. The ability to detect plant diseases early is not just a step forward in agricultural technology; it is a leap towards securing the future of food production worldwide.

II. LITERATURE REVIEW
[1]In the investigation led by Benjamín Luna Benoso, José Cruz Martínez-Perales, Jorge Cortés-Galicia, Rolando Flores-Carapia, and Víctor Manuel Silva-García on the detection of diseases in tomato leaves through color analysis and GLCM methodology, the study identifies a challenge marked by low accuracy in effectively distinguishing between healthy and diseased leaves.
[2]In the work conducted by Mohammed Brahimi, Kamek Boukhalfa, and Abdelouahab Moussaoui on the classification and visualization of symptoms of tomato diseases, challenges are evident with low accuracy and limited feature extraction. The study explores the application of neural networks (NN, ANN) and support vector machines (SVM) to address these issues and enhance the efficiency of disease classification in tomato plants.
[3]In the research conducted by H. Al-Hiary, et al on the detection and classification of plant diseases, decision trees, SVM, and K-means algorithms are employed. [4]The study notes a challenge with a higher dependency on hyperparameters, emphasizing the need for careful parameter tuning to optimize the performance.
[5]Nishanth Vijay's study on the detection of plant diseases in tomato leaves using CNN and KNN highlights a constraint related to limited temporal information, indicating a potential challenge in capturing the temporal aspects of disease progression in the analyzed leaves.
[6]In the research conducted by Jinpeng Chen et al on the application of a CNN-LSTM hybrid network for Intrusion Detection System (DL-IDS), it is revealed that the model experiences low detection accuracy specifically concerning Heartbleed attacks due to insufficient data availability.[7] This emphasizes the significance of addressing data scarcity challenges for more robust intrusion detection systems.
[8]Abdulkadir Tasdele and Baha Sen propose a hybrid CNN-LSTM model for the classification of pre-miRNAs. However, the study notes a limitation attributed to a restricted dataset, emphasizing the need for further data acquisition to enhance the model's classification capabilities in pre-miRNA analysis.
[9]Melike Sardogan, Adem Tuncer et al contribute to plant leaf disease detection and classification using a combination of CNN and the Learning Vector Quantization (LVQ) algorithm. However, the study highlights challenges with low accuracy and limited feature extraction capabilities, suggesting a need for further refinement in the model for more effective disease identification.
[10]V. Sunanthini, J. Deny et al undertake a study comparing various CNN algorithms for feature extraction on fundus images with the objective of detecting glaucoma. This research focuses on evaluating the performance and effectiveness of different CNN architectures in the glaucoma diagnosis based on fundus image analysis. [11]Rajath Ramakrishna explores a machine learning-based approach employing XGBoost and VGG16 models. However, the study acknowledges a limitation in being restricted to static images, suggesting potential challenges in capturing dynamic aspects of disease progression in real-time or temporal sequences.
[12]In Aishwarya V's work on tomato leaf disease detection and classification using CNN, the model demonstrates strong performance but occasionally exhibits overfitting issues, emphasizing the need for regularization techniques or architectural adjustments to enhance the model's generalization capabilities.
[13In the research conducted by Seshaiah Merilkapudi on employing Long Short-Term Memory (LSTM) for early forest fire smoke recognition, a challenge arises due to data imbalance. This highlights the importance of dealing with class imbalance issues to ensure the robustness and accuracy of the LSTM model in detecting forest fire smoke in its early stages.
[14]Poormehdi Ghaemmagham's study on "Tracking of Humans in Video Stream Using LSTM Recurrent Neural Network" contributes by exploring the capabilities of LSTM networks in accurately tracking human subjects within video streams.[15]A critical aspect addressed in the literature review is the paramount importance of data quality and quantity in training LSTM models effectively. It underscores the challenges posed by poor-quality data.
III. METHODOLOGY
A. CNN-LSTM
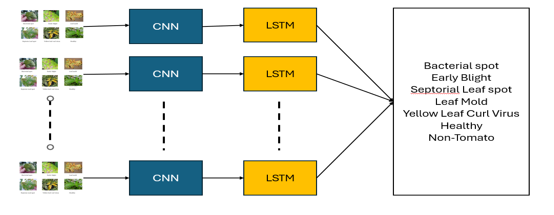
The methodology centers on employing a CNN-LSTM hybrid model for tomato leaf disease detection. This approach integrates Convolutional Neural Networks (CNNs) and Long Short-Term Memory (LSTM) networks to effectively capture spatial features from images and model temporal dependencies in sequential data, respectively. The CNN-LSTM architecture enables the extraction of intricate spatial patterns from the provided images using convolutional layers, followed by the utilization of LSTM layers to learn temporal relationships between the extracted features.
By combining the advantages of both CNNs and LSTMs, the model can effectively analyze the complex patterns present in the tomato leaf images, leading to accurate disease detection. This methodology underscores the significance of leveraging hybrid deep learning architectures for addressing challenges in agricultural disease detection tasks.
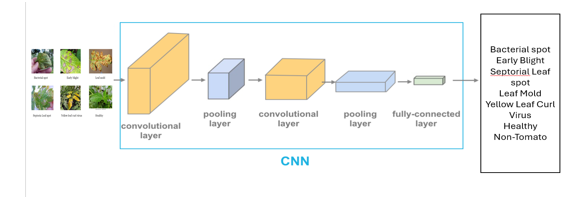
- Convolutional Neural Networks (CNN)
Convolutional Neural Networks are a class of deep neural networks, highly effective for handling data with a grid-like structure, such as images. CNNs are particularly powerful for tasks like image recognition, image classification, object detection, and many others within computer vision fields.
2. Core Components of CNNs
a. Convolutional Layers: The primary building blocks of a CNN. These layers perform convolution operations, applying filters (or kernels) to the input data to create feature maps. This process helps the network learn specific features of the input data, such as edges in an image, by focusing on small regions of input data at a time.
b. ReLU (Rectified Linear Unit) Layer: Often follows a convolutional layer; it introduces non-linearity into the model, allowing it to learn more complex patterns. The ReLU function typically transforms all negative values in the feature maps to zero, keeping the positive values unchanged.
c. Pooling Layers: These layers reduce the spatial dimensions of the input volume for the next convolutional layer. Max pooling reduces dimensionality by keeping the maximum value from each cluster of inputs while discarding the rest. This operation not only reduces computational load and memory usage but also helps with making the detection of features somewhat invariant to scale and orientation changes.
d. Fully Connected (FC) Layers: After multiple convolutional and pooling layers, the high-level reasoning in the neural network is done through FC layers. In a fully connected layer, neurons are connected to all activations in the preceding layer, similar to conventional neural networks. Their role is to output predictions from the network.
e. Softmax or Sigmoid Layer: In classification tasks, the final fully connected layer is often subsequently accompanied by a softmax or sigmoid activation function that maps the output into a probability distribution over predefined classes (in case of softmax) or a binary outcome (in case of sigmoid).
B. Long Short-Term Memory (LSTM)
Long Short-Term Memory (LSTM) networks are a special kind of Recurrent Neural Network (RNN) capable of learning longterm dependencies. They were implemented to tackle the issue of the vanishing gradient problem that affects standard RNNs. LSTMs are especially beneficial for tasks involving sequence prediction because they can retain information over extended durations, making them ideal for applications in natural language processing, time series analysis, and more.
- Core Components of LSTM
a. Forget Gate Operation: The forget gate looks at ht-1 and xt and decides what information are we discarding from the cell state.
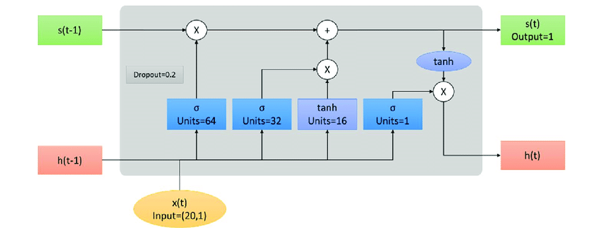
b. Input Gate Operation: Simultaneously, the input gate determines which values within the cell state to update with new information generated by tanh layer
c. Cell State Update: The old cell state, Ct-1 and is updated into the new cell state, Ct. The operations from the forget and input gates are applied to discard or update information.
d. Output Gate Operation: Finally, the output gate decides what the next hidden state ht should be. This hidden state holds data on earlier inputs and is utilized for predictions or as input to the next LSTM cell in the sequence.
2. Implementation
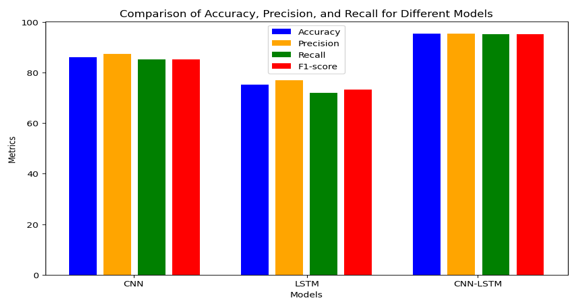
The introductory phase of our research project involved the exploration and evaluation of three distinct models: Convolutional Neural Network (CNN), Long Short-Term Memory (LSTM), and a hybrid CNN-LSTM architecture. These models were selected for their potential to accurately detect and categorize diseases affecting tomato leaves based on image data. Our primary objective was to compare the performance of these models in terms of accuracy and other relevant metrics, aiming to ascertain the most suitable approach for the task at hand.
3. Data preprocessing
Initially, the focus was on data cleaning and transformation, utilizing the OpenCV library for image processing and NumPy for data manipulation. The dataset comprised images of healthy and infected tomato leaves categorized into seven classes including Bacterial Spot, Early Blight, Septoria Leaf Spot, Yellow Leaf Curl Virus, Leaf Mold, Healthy leaf, Non-Tomato leaf.The dataset was meticulously split into training and testing sets, ensuring a balanced representation of each class in both sets. One-hot encoding was applied to facilitate model training, converting categorical labels input data, reducing into binary vectors for effective neural network learning from categorical data.
D. CNN-LSTM

The CNN-LSTM hybrid model was constructed using the TensorFlow Keras API. The model architecture included convolutional layers for feature extraction, followed by LSTM layers for sequence learning. Rectified Linear Unit (ReLU) activation functions were employed for the convolutional layers, while Softmax activation was used for the output layer to generate probability distributions over the disease classes.
To mitigate overfitting, dropout regularization was incorporated after each convolutional layer, and max-pooling layers were utilized to downsample the spatial dimensions of the computational complexity while retaining critical features.
During the training phase, the model was compiled using the Adam optimizer and categorical cross-entropy loss function. Training was conducted over multiple epochs, with adjustments made to hyperparameters such as batch size and learning rate to optimize performance. Validation data were used to monitor the training process and evaluate model metrics, including accuracy, loss, precision, and recall.
D. CNN
The CNN architecture comprised multiple layers, including convolutional, max-pooling, dropout, and dense layers. The convolutional layers were tasked with extracting crucial features from the input images, whereas the max-pooling layers enabled spatial down-sampling to decrease computational complexity. Dropout layers were integrated to combat overfitting by randomly deactivating a portion of neurons during training. The final dense layer employed a softmax activation function to produce a probability distribution over the possible output classes. The model was compiled using the Adam optimizer and categorical cross-entropy loss function, with accuracy as the primary evaluation metric.
D. LSTM
To facilitate model training, the target labels underwent one-hot encoding, ensuring compatibility with the LSTM's categorical output. Concurrently, the input data underwent reshaping to conform to the LSTM layer's expected input shape. This reshaping operation transitioned the input data into a 3D array format, featuring dimensions indicative of the dataset's sample count, the number of frames in each sequence (time_steps), and the flattened size of each frame (features).
The LSTM model architecture comprised a singular LSTM layer outfitted with 100 units, followed by a dense layer leveraging softmax activation to yield output probabilities for each class. Subsequently, the model underwent compilation utilizing the Adam optimizer and categorical cross-entropy loss function, with accuracy serving as the primary evaluation metric. The LSTM model was subsequently trained on the reshaped training dataset. Throughout the training process, the model adeptly assimilated temporal dependencies within input sequences, enabling it to make accurate predictions regarding the presence of various tomato leaf diseases.

V. LIMITATIONS
One limitation of our study is the potential imbalance in the distribution of classes within the dataset. Imbalanced datasets, where certain classes are overrepresented or underrepresented compared to others, can bias the model towards the majority classes and lead to reduced performance on minority classes. In the case of tomato leaf disease detection, some diseases may occur more frequently than others in agricultural settings, resulting in unequal class distributions in the dataset. This imbalance can affect the model's ability to accurately identify fewer common diseases, thereby reducing the overall effectiveness of the system. To address this limitation, techniques such as oversampling, under sampling, or the use of class weights during training can be employed to ensure equal representation of all classes and improve the model's performance across the entire spectrum of diseases.
VI. FUTURE WORK
Expanding the scope of diseases covered in our research offers a hopeful guidance for forthcoming. While our current model focuses on detecting five common tomato leaf diseases, there are numerous other diseases that pose significant threats to tomato crops worldwide. By incorporating additional diseases into our detection system, we can provide farmers with a more comprehensive tool for identifying and managing a broader range of plant health issues. This expansion would require the collection of extensive datasets encompassing various diseases, as well as the development of robust classification models capable of accurately distinguishing between different disease types.
Moreover, addressing a wider array of diseases would enhance the versatility and applicability of our technology, ultimately contributing to more effective disease management strategies.
VII. ACKNOWLEDGEMENTS
We are grateful to our project Guide, Dr. Ramchandran Sir, and project Coordinator, Sri Hari Sir, for their important direction, support, and insightful input during the project's development. Their experience and encouragement has helped shape our strategy and overcome obstacles. Special thanks are due to the creators and curators of publicly available datasets, without which our research efforts would not have been feasible.
We appreciate the work of people who gathered, annotated, and shared these excellent resources, which serve as the foundation for training and assessing deep learning models for disease detection. e also owe gratitude to the participants who provided valuable insights and feedback during the course of this study. Lastly, we acknowledge the efforts of the entire research team for their dedication and hard work in conducting this research.
Conclusion
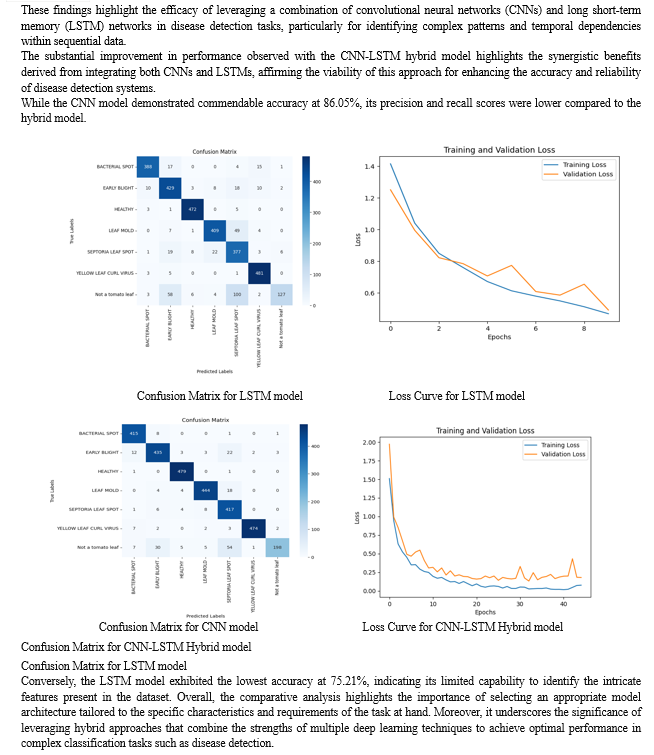
The research conducted offers a thorough investigation into the efficacy of convolutional neural networks (CNNs), long short-term memory networks (LSTMs), and their hybrid, CNN-LSTM, in the realm of tomato leaf disease detection. Through experimentation and analysis, we have showed that the CNN-LSTM hybrid model outperforms both individual models, achieving an impressive accuracy of 95.33%. This underscores the potential of combining CNNs for feature extraction with LSTMs for temporal information processing in disease detection tasks. Furthermore, our results emphasize the importance of leveraging deep learning techniques in agriculture, offering a promising avenue for enhancing food security and sustainability. Our study contributes to the field by precisely identifying and categorizing diseases affecting tomato leaves empowering smallholder farmers with timely and effective disease management strategies. Moving forward, continued research in this area holds promise for further advancements in crop health monitoring and agricultural productivity.
References
[1] Luna-Benoso, Benjamín, José Cruz Martínez-Perales, Jorge Cortés-Galicia, Rolando Flores-Carapia, and Víctor Manuel Silva-García. \\\"Detection of diseases in tomato leaves by color analysis.\\\" Electronics 10, no. 9 (2021): 1055. [2] Brahimi, Mohammed, Kamel Boukhalfa, and Abdelouahab Moussaoui. \\\"Deep learning for tomato diseases: classification and symptoms visualization.\\\" Applied Artificial Intelligence 31, no. 4 (2017): 299-315. [3] Al-Hiary, Heba et al \\\"Fast and accurate detection and classification of plant diseases.\\\" International Journal of Computer Applications 17, no. 1 (2011): 31-38. [4] Vijay, Nishanth. \\\"Detection of plant diseases in tomato leaves: with focus on providing explainability and evaluating user trust.\\\" (2021). [5] Sun, Pengfei et al.\\\"DL-IDS: Extracting features using CNN-LSTM hybrid network for intrusion detection system.\\\" Security and communication networks 2020 (2020): 1-11. [6] Tasdelen, Abdulkadir, and Baha Sen. \\\"A hybrid CNN-LSTM model for pre-miRNA classification.\\\" Scientific reports 11, no. 1 (2021): 14125. [7] Sardogan, Melike, Adem Tuncer, and Yunus Ozen. \\\"Plant leaf disease detection and classification based on CNN with LVQ algorithm.\\\" 382-385. IEEE, 2018. [8] Sunanthini, V., J. Deny, S. Vairaprakash, Petchinathan Govindan, S. Sudha, V. Muneeswaran, and M. Thilagaraj. \\\"Comparison of CNN algorithms for feature extraction on fundus images to detect glaucoma.\\\" [9] Chowdhury, M. E., Tawsifur Rahman et al. \\\"Tomato leaf diseases detection using deep learning technique.\\\" Technology in Agriculture 453 (2021) [10] Pushpa, B. R., and V. V. Aiswarya. \\\"Tomato leaf disease detection and classification using CNN.\\\" 71, no. 4 (2022): 2921-2930. [11] Sathishkumar, Veerappampalayam et al. \\\"Forest fire and smoke detection using deep learning-based learning without forgetting.\\\" Fire ecology 19, no. 1 (2023): 9. [12] Ghaemmaghami, Masoumeh P. \\\"Tracking of humans in video stream Using LSTM recurrent neural network.\\\" Master in Machine Learning, School of Computer Science And Communication, KTH Royal School of Computer Science And Communication (2019). [13] Sengan S et al mathematical analysis for comprehensive real-time data-driven in healthcare. Math. Eng. Sci. Aerosp. (MESA) 2021;12:77–94. [14] Wisesa O., Andriansyah A., Khalaf O.I. Prediction Analysis for Business to Business (B2B) Sales of Telecommunication Services using Machine Learning Techniques. Majlesi J. Electr. Eng. 2020;14:145–153. doi: 10.29252/mjee.14.4.145. [15] Basic study of automated diagnosis of viral plant diseases using convolutional neural networks [16] Sankaran, S. et al. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 72(1), 1–13 (2010). [17] D. Das, M. Singh, S. S. Mohanty, S. Chakravarty (2020), Leaf Disease Detection Using Support Vector Machine, (ICCSP) (pp 1036-1040) [18] Alex Krizhevsky, Ilya Sutskever and Geoffrey E. Hinton, \\\"Imagenet classification with deep convolutional neural networks\\\", Advances in neural information processing systems, pp. 1097-1105, 2012. [19] He, Y. Z., Wang, Y. M., Yin, T. Y., Fiallo-Olivé, E., Liu, Y. Q., Hanley-Bowdoin, L., et al. (2020). A plant DNA virus replicates in the salivary glands of its insect vector via recruitment of host DNA synthesis machinery. Proc. Natl. Acad. Sci. United States America 117 (29), 16928–16937. doi: 10.1073/pnas.1820132117 [20] Valenzuela, M. E. M. & Restovi?, F. Valorization of tomato waste for energy production. In Tomato Chemistry, Industrial Processing and Product Development, 245–258 (2019).
Copyright
Copyright © 2024 Dr. V. Rama Chandran, M Jyothika, M Nandhini, P. Neelima, P Neeraja. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET59173
Publish Date : 2024-03-19
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

